Figure 6.
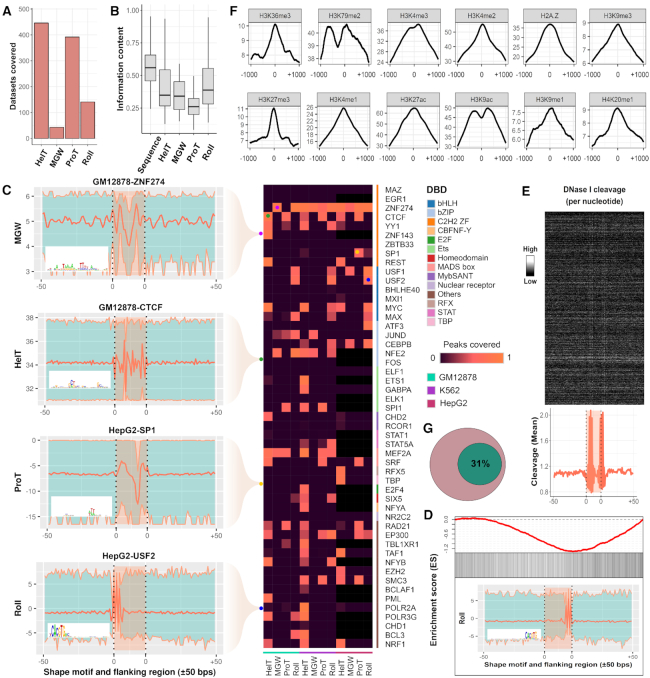
A comprehensive analysis of the identified shape motifs. (A) The number of datasets in the 690 ENCODE ChIP-seq datasets that were covered by shape motifs of HelT (446), MGW (43), ProT (392) and Roll (141). (B) IC of underlying sequences of the identified sequence motifs and shape motifs. (C) Each entry in the heatmap indicates the ratio of peaks covered by the four kinds of shape motifs which were identified in the 51 TFs within GM12878, K562 and HepG2 cell lines, while the black entries represent missing values. Four representative shape motif logos were listed at the left side, where each of them represents the shape motif profile and 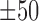 bps flanking regions using a bold orange curve. The two boundary curves of the blue region represent upper and lower bounds of shape features in the corresponding motif instances. (D) The enrichment score of MAX’s Roll motif in its corresponding ChIP-seq peaks in the K562 cell line. Black ticks indicate the occurrence of ChIP-seq peaks that contain at least one instances of MAX’s Roll motif. (E) The heat map of per-nucleotide DNase I cleavage of TFBSs of MAX’s Roll as well as
bps flanking regions using a bold orange curve. The two boundary curves of the blue region represent upper and lower bounds of shape features in the corresponding motif instances. (D) The enrichment score of MAX’s Roll motif in its corresponding ChIP-seq peaks in the K562 cell line. Black ticks indicate the occurrence of ChIP-seq peaks that contain at least one instances of MAX’s Roll motif. (E) The heat map of per-nucleotide DNase I cleavage of TFBSs of MAX’s Roll as well as 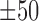 bps flanking regions, where each row represents a motif instance. The orange curve represents mean DNase I cleavage. (F) Twelve histone marks of
bps flanking regions, where each row represents a motif instance. The orange curve represents mean DNase I cleavage. (F) Twelve histone marks of 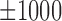 bps around the summits of Max's Roll motif. (G) The average ration (31%) of ChIP-seq peaks that cannot be explained by the 100 TFs within the K562 cell line.
bps around the summits of Max's Roll motif. (G) The average ration (31%) of ChIP-seq peaks that cannot be explained by the 100 TFs within the K562 cell line.