Figure 2.
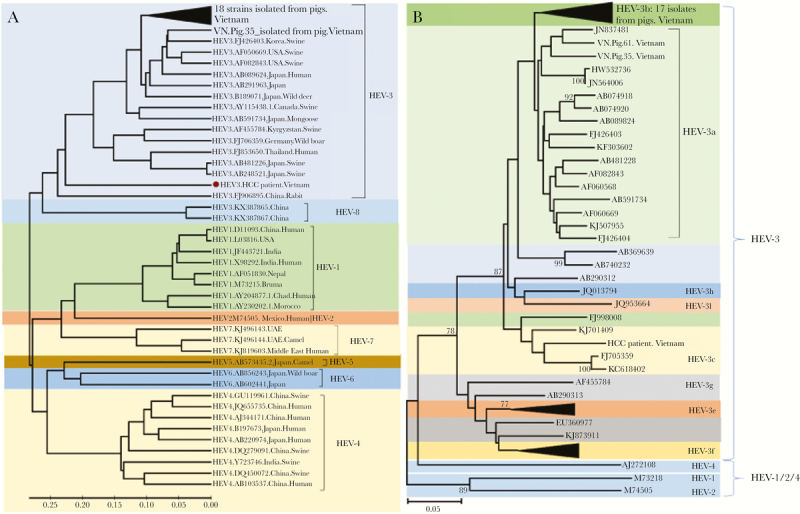
Phylogenetic analysis of 19 hepatitis E virus (HEV) strains isolated from domestic pigs. (A) Phylogenetic tree was constructed based on the alignment of 306 base pairs of the HEV RNA-dependent RNA polymerase (RdRp) region (ORF1) of 19 nucleotide sequences isolated from domestic pigs, 1 HEV strain isolated from a liver cancer patient coinfected with hepatitis B virus in Vietnam. Forty full-length HEV genomes isolated from animals and humans (HEV-1 to HEV-8) retrieved from the NCBI database along with GenBank accession numbers were included in the analysis. A neighbor-joining tree was constructed with a bootstrap of 1000 replicates. Genetic distances that are in the units of the number of base substitutions per site were computed using the Kimura-parameter method are given. The bar at the base of the tree indicates the scale for nucleotide substitutions per position. (B) Phylogenetic tree constructed for all identified HEV genotype 3 sequences. The analysis involved 34 nucleotide sequences, including 19 HEV strains isolated from domestic pigs in Vietnam. Bootstrap analysis values (percentages) are shown.
