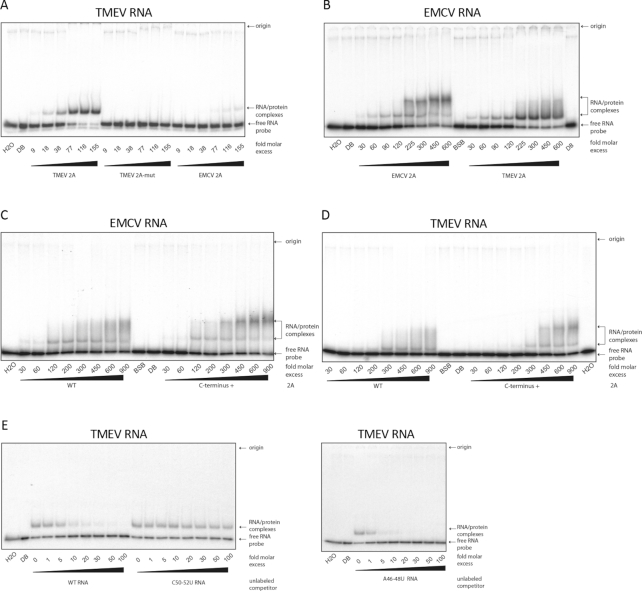
Figure 8.
EMSA analysis of cardiovirus 2A RNA binding activity. (A) 32P-labelled RNA containing the WT TMEV PRF signal was incubated with increasing amounts of TMEV 2A, TMEV 2A-mut, or EMCV 2A and subjected to EMSA on 4% non-denaturing acrylamide gels. (B) 32P-labelled RNA containing the WT EMCV PRF signal was incubated with increasing amounts of EMCV 2A or TMEV 2A. (C) 32P-labelled RNA containing the WT EMCV PRF signal was incubated with increasing amounts of EMCV 2A (WT) or the C-terminally extended version of EMCV 2A (C-terminus +). (D) 32P-labelled RNA containing the WT TMEV PRF signal was incubated with increasing amounts of TMEV 2A (WT) or the C-terminally extended version of TMEV 2A (C-terminus +). (E) Unlabelled competitor RNA containing the WT, C50–52U mutant or A46–48U mutant TMEV PRF signal was incubated with 32P-labelled RNA containing the WT TMEV PRF signal, and TMEV 2A (0.7 μM), and analyzed by EMSA. In (A, B, E), WT and all mutant 2As were the C-terminally extended versions (see Methods). In (A–D), numbers below lanes show fold molar excess of 2A with respect to RNA (10 nM). In (E), numbers below lanes show fold molar excess of competitor RNA with respect to 32P-labelled WT RNA (10 nM). In lanes BSB, DB and H2O, RNA was incubated alone with band-shift buffer, protein dilution buffer or water, respectively.