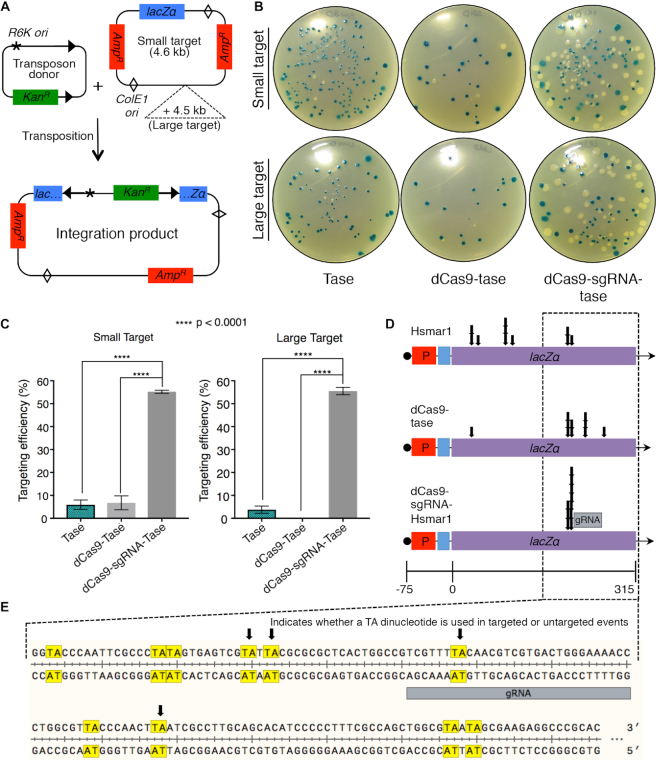
Figure 5.
Transposon targeting reactions. (A) Illustration of the transposon donor and target plasmids. The donor plasmid (pRC704) encodes an Hsmar1 transposon with an R6K origin of replication and a kanamycin resistance gene. The target plasmid (pRC2312) is essentially a dimer of pBluescript with one copy of the lacZα gene removed. Since the plasmid has two origins and to ampicillin markers it can tolerate transposon insertions anywhere. The large target has an extra 4.5 kb of non-specific DNA. Reactions also contained a background of non-specific DNA. (B) Reactions were performed with the indicated transposase proteins, transformed into Escherichia coli and plated on LB plus ampicillin, kanamycin and X-gal. Transposon integrations into the lacZα gene yield white colonies. Integrations elsewhere yield lacZ+ colonies. (C) Targeting efficiency. Error-bars are standard error of the mean where n = 6 biological replicates. Ordinary one-way ANOVA analysis: Small target; transposase versus dCas9-transposase, P = 0.95; transposase versus dCas9-sgRNA-transposase, P = <0.0001; dCas9-transposase versus dCas9-sgRNA-transposase, P = <0.0001. Large target: transposase versus dCas9-transposase, P = 0.14; transposase versus dCas9-sgRNA-transposase, P = <0.0001; dCas9-transposase versus dCas9-sgRNA-transposase, P = <0.0001. (D) Ten white colonies were picked from the three sets of plates and the location of the transposon insertion determined by Sanger sequencing. (E) An expanded view of the indicated area of the lacZα gene.