Figure 4.
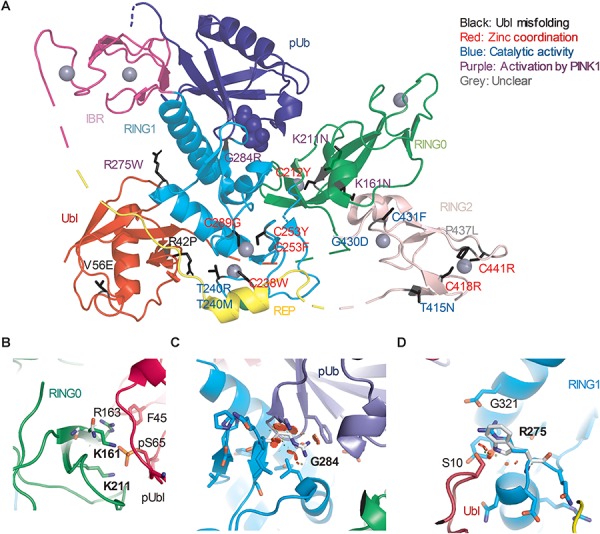
Structural analysis of Parkin variants revealed various pathogenic mechanisms. (A) Pathogenic variants were mapped onto the 3D structure of human Parkin bound to pUb (PDB 5N2W). The side chains of the amino acids substituted by the variants were highlighted in black. The blue spheres represent the phosphate of pUb. The gray spheres represent zinc. The color of the text indicates the type of disruption to Parkin caused by the variant. (B) Close-up view of pUbl-RING0 interface in the structure of fly pParkin bound to phospho-Ub (PDB 6DJX). Lys161 and Lys211 form ionic interactions with the phosphate on Ser65 of the pUbl. Mutations of these lysine residues would weaken the pUbl-RING0 interaction, preventing activation of Parkin. (C) Close-up view of the pUb:RING1 interface in human Parkin bound to pUb (PDB 5N2W). The G284R variant in Parkin RING1 would introduce major clashes with pUb, disrupting the interaction. (D) Close-up view of Arg275 in human Parkin bound to pUb (PDB 5N2W). Arg275 interacts with Glu321 in the helix that interacts with pUb. Mutation to a tryptophan (white) would introduce clashes with this helix as well as Ser10 in the Ubl domain.
