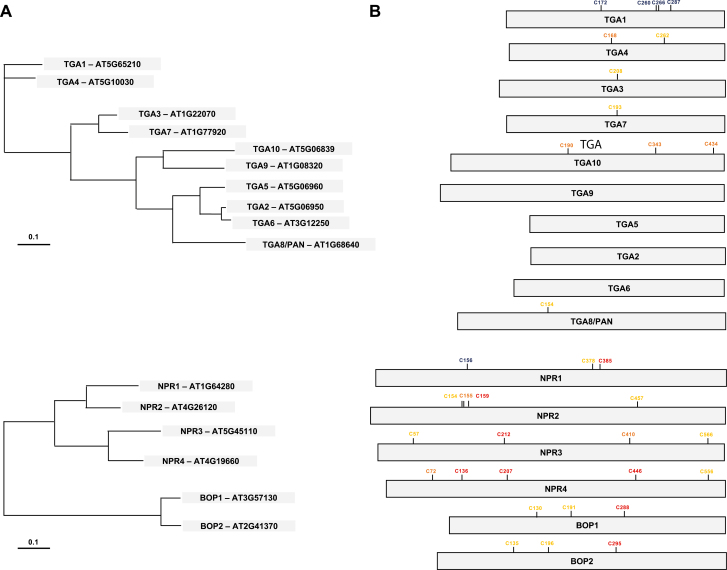
Fig. 4.
S-Nitrosation analysis in group D of bZIP transcription factor and NONEXPRESSOR OF PATHOGENESIS-RELATED GENES (NPR) families. (A) Dendrograms of TGA members of the group D bZIP transcription factor family and NPR-like proteins. The branch length is proportional to the number of substitutions per site (http://phylogeny.lirmm.fr/). (B) In silico prediction of S-nitrosation Cys (C) targets by using the GPS-SNO 1.0 software (Xue et al., 2010). The analysis shows target Cys in red, orange, and yellow depending on the S-nitrosation score (high, medium, and low, respectively). The Cys residues highlighted in blue correspond to in vivo and/or in vitro S-nitrosation.