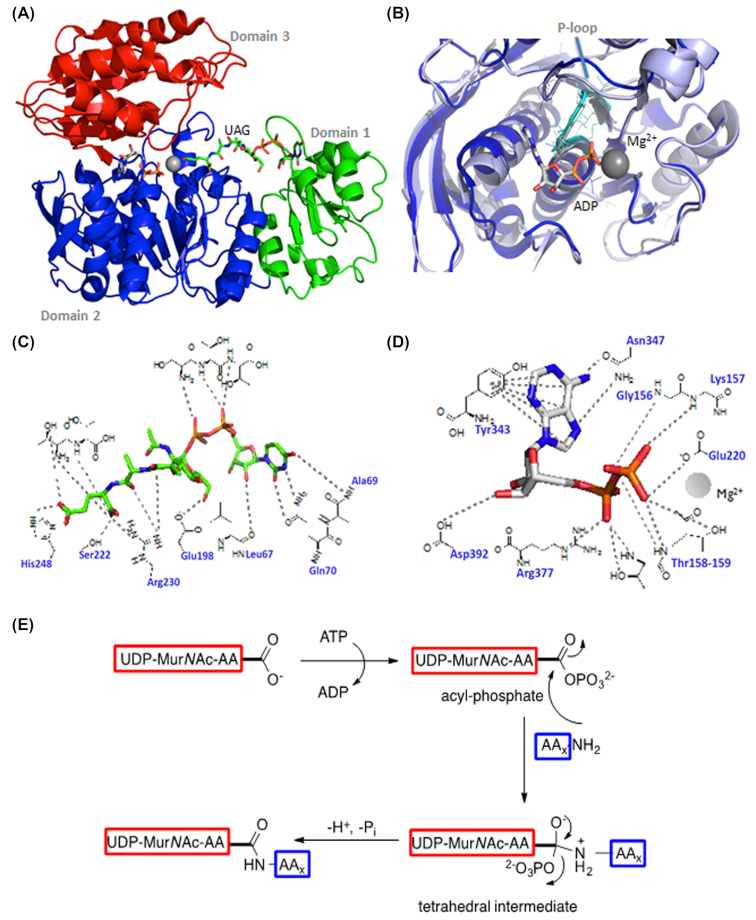
Figure 3.
(A), Cartoon representation of the crystal structure of MurE from M. tuberculosis. The UDP substrate binding domain 1 is shown in green, the central ATPase domain 2 is shown in blue and the amino acid substrate binding domain 3 is shown in red. Image generated using PyMOL (PDB code: 2XJA). (B), Overlay of domain 2 from MurE from E. coli (PDB code: 1E8C, light blue) and M. tuberculosis (PDB code: 2XJA, dark blue). The P-loops are highlighted in cyan. (C), Interaction map of the UDP-MurNAc substrate with MurE from M. tuberculosis. (D), Interaction map of ADP with MurE from M. tuberculosis. (E), The reaction mechanism of Mur ligases. The ‘AA’ in the UDP-MurNAc-AA substrate represents the peptide stems of varying lengths whereas the AAx.NH2 represents the incoming amino acid.