2.
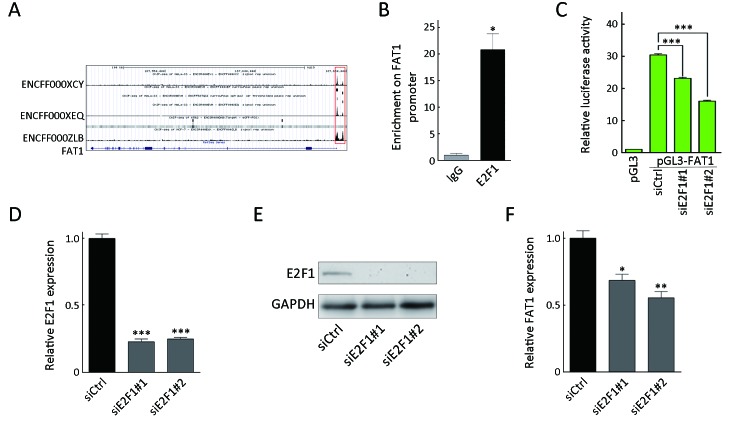
E2F1 regulates FAT1 transcription. (A) Putative binding sites of E2F1 on the FAT1 promoter region from different cohorts in encyclopedia of DNA elements (ENCODE) database. Red box represents binding reads of E2F1; (B) Enrichment of E2F1 on FAT1 promoter in KYSE30 cells shown by chromatin immunoprecipitation (ChIP) and quantitative polymerase chain reaction (qPCR). Enrichment is determined as the amount of FAT1 promoter associated to E2F1 relative to immunoglobulin G (IgG) control; (C) Luciferase activity of pGL3-FAT1 vector was measured in KYSE30 cells upon E2F1 knockdown. Data are presented as ratio of the firefly luciferase activity to Renilla luciferase activity; RT-qPCR (D) and Western blot (E) analyses of E2F1 expression in KYSE30 cells upon E2F1 knockdown. Relative RNA levels of E2F1 were normalized to endogenous GAPDH; (F) RT-qPCR analysis of FAT1 expression in KYSE30 cells as described in (D). Relative RNA levels of FAT1 were normalized to endogenous GAPDH. *, P<0.05; **, P<0.01; ***, P<0.001vs. control.