Figure 1.
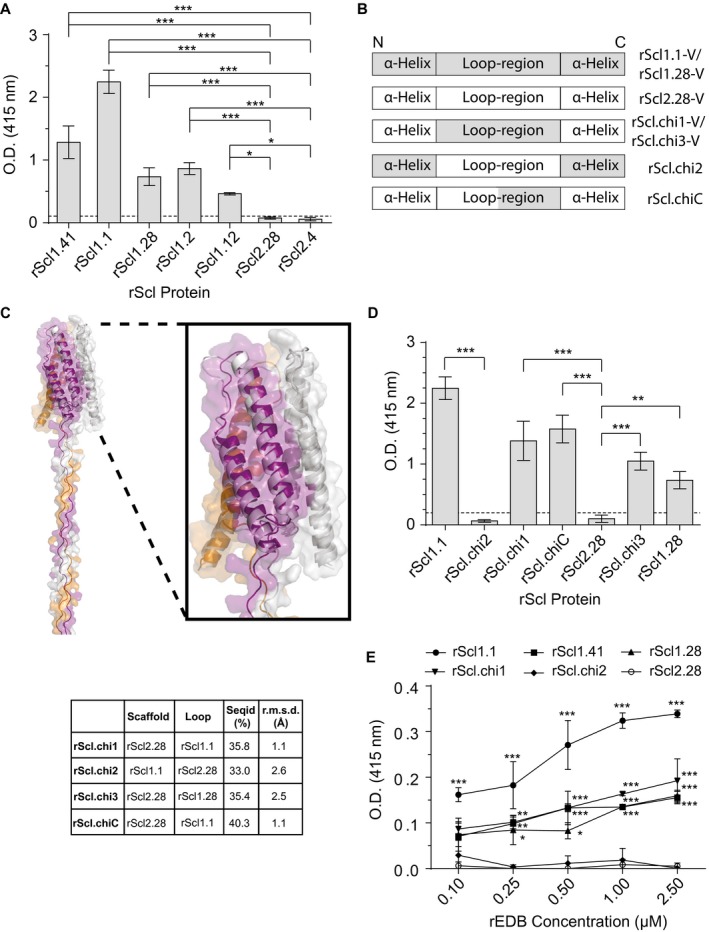
Scl1‐V domain binds fibronectin type III repeat, extra domain B (EDB), via surface‐exposed loops. Recombinant extra domain B (rEDB) was tested for binding to recombinant streptococcal collagen‐like proteins (rScl). A. rEDB binding to Scl1‐ and Scl2‐derived rScl constructs. rScl proteins were immobilized onto Strep‐Tactin‐coated microplate wells and incubated with rEDB. Primary anti‐His‐tag mAb and HRP‐conjugated secondary Ab were used for ligand detection by ELISA. Graph bars indicate the mean OD415nm normalized against BSA controls. Statistical analysis was calculated using a one‐way ANOVA, from three independent experiments, each performed in triplicate wells (N = 3 ± SD); *P ≤ 0.05, ***P ≤ 0.001. Statistical significance evaluates the differences in rEDB binding by rScl1 proteins, as compared to ECM‐binding negative rScl2.28 and rScl2.4 control proteins. Dashed line indicates threshold OD415nm +2SD values recorded for binding‐negative rScl2.28 control protein. B. Schematic representation of the variable (V) domains in recombinant Scl constructs used. Homotrimeric rScl1.1‐ and rScl1.28‐V domains (gray box), and rScl2.28‐V domain (white box) each consists of three conserved pairs of anti‐parallel α‐helices, with interconnecting loops (McNitt et al., 2018). Chimeric proteins were generated by replacing either the entire (rScl.chi1‐3) or partial (rScl.chiC) loop sequences between different constructs. C. I‐TASSER modeling of chimeric rScl proteins. Left, far‐out view of a representative I‐TASSER model of Scl.chi1, including the V domain and the first 16 triplets of the CL domain. Right insert, close‐up view of the Scl.chi1‐V domain. The three monomers are colored purple, orange and gray in both models. In close‐up view, white depicts Scl2.28, the loop‐host Scl protein of Scl.chi1. Bottom, I‐TASSER model sequence identities and root mean square deviations from Scl2.3, room mean square deviations performed using DALI server. D. rEDB binding to chimeric rScl constructs. ELISA was performed as described in panel A. Statistical analysis was calculated using a one‐way ANOVA, from three independent experiments, each performed in triplicate wells (N = 3 ± SD); **P ≤ 0.01, ***P ≤ 0.001. Statistical significance evaluates the differences in rEDB binding between chimeric proteins and their respective loop‐hosts: rScl.chi1, rScl.chiC and rScl.chi3 compared to binding‐negative control protein rScl2.28, and rScl.chi2 to binding‐positive control protein rScl1.1. Dashed line indicates threshold OD415nm +2SD values recorded for binding‐negative rScl2.28 control protein. E. Concentration‐dependent binding of rEDB to rScl proteins. rScl proteins, immobilized onto Strep‐Tactin coated microplate wells, were incubated with increasing concentrations of rEDB (0.1–2.5 µM) and detected by ELISA, as described above. Statistical analysis was calculated using a two‐way ANOVA, from three independent experiments, each performed in triplicate wells (N = 3 ± SD); *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Statistical significance evaluates the differences in rEDB binding by rScl1 proteins, rScl.chi1 and rScl.chi2, as compared with ECM‐binding negative rScl2.28 control protein. [Colour figure can be viewed at wileyonlinelibrary.com]
