Fig. 2. The citrate-binding pocket and the effect of mutations in the binding pocket on chemotaxis to citrate.
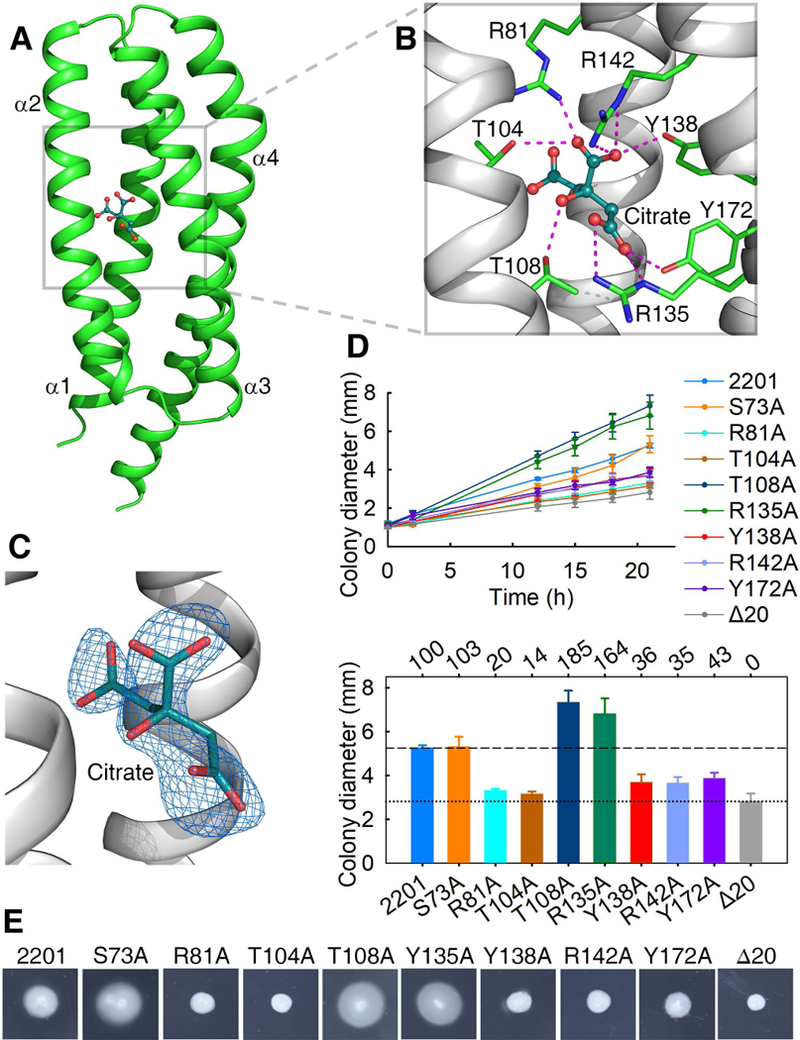
(A) The structure of a citrate-bound monomer. (B) The ligand-binding pocket. Residues involved in ligand-binding and the ligand (citric acid) are shown in stick and ball-and-stick models, respectively. (C) Citrate is surrounded by ligand-omit 1FoFc electron density map contoured at 3σ. (D) The plots of swimming ring diameter vs time for strains harboring MCP2201 mutants with substitutions in amino acid residues at the ligand binding pocket are shown in the upper panel. Averages of colony diameters of site-directed mutants were plotted, and standard deviations are shown, n=3. The relative chemotaxis abilities of mutants are shown in the bottom panel, with percentage labelled above histogram of panel. The levels of chemotactic response of chemotaxis-null mutant CNB-1Δ20 harboring or not wild-type MCP2201 towards citrate are normalized as 100% and 0%. Mutant S73A was neutral to chemotaxis and was chosen as a control. (E) One set of typical motility plate assay results for MCP2201 mutants.
