Fig. 3. The citrate-bound LBD trimer-of-monomers structure.
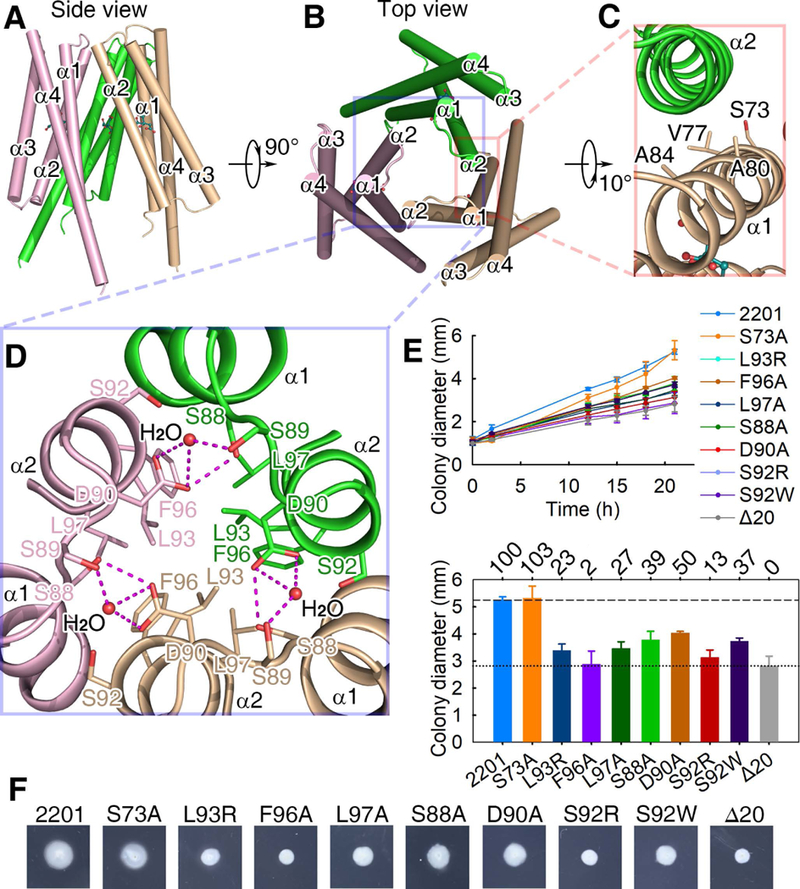
(A, B) The side (A) and top (B) views. (C) The interface between two monomers of the trimeric structure. (D) The three-fold symmetric interface of the trimeric structure. Residues L93, F96 and L97 form a hydrophobic patch and then interact with their counterparts in the other protomers. The distance between L97 of one protomer and F96 of another protomer is 4.2 Å. (E) The plots of swimming ring diameter vs time for strains harboring MCP2201 mutants. Substitutions in amino acid residues at the trimeric interface are shown in the upper panel, whereas the relative chemotaxis abilities of mutants are shown in the bottom panel with percentage labelled above histogram of panel. The standard deviations are shown, n=3. (f) One set of typical motility plate assay results for MCP2201 mutants.
