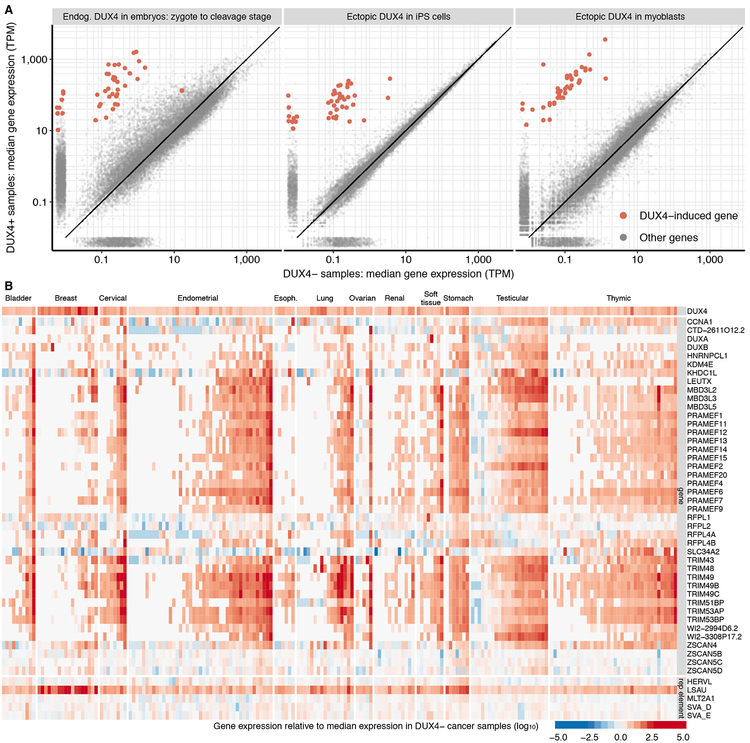
Figure 3. DUX4 drives an embryonic gene expression program in cancer.
(A) Differential gene expression induced by endogenous or ectopic DUX4. Each point represents mRNA levels for a single gene in samples with (y axis) or without (x axis) DUX4 expression. Plots illustrate comparisons between cleavage-stage embryos and zygotes, which have high and low DUX4 expression (left panel), iPSCs with or without DUX4 induction (center panel), and myoblasts with or without DUX4 induction (right panel) (Feng et al., 2015; Hendrickson et al., 2017). Red, high-confidence list of DUX4-induced genes identified by intersecting the sets of up-regulated genes in all three illustrated comparisons. TPM, transcripts per million.
(B) Expression of high-confidence DUX4 targets in DUX4+ cancers. Heat map illustrates the expression of each DUX4 target (rows) in each individual DUX4+ sample (columns) relative to the median expression across all DUX4− samples from that cancer type. The LSAU repetitive element corresponds to the beta satellite repeat, which is a multicopy genomic element like DUX4. Approximately 55% of LSAU’s 2,759 nt consensus sequence overlaps with part of DUX4’s consensus sequence, so its expression is invariably higher in DUX4+ samples when expression is quantified via short-read sequencing.
See also Figure S3.