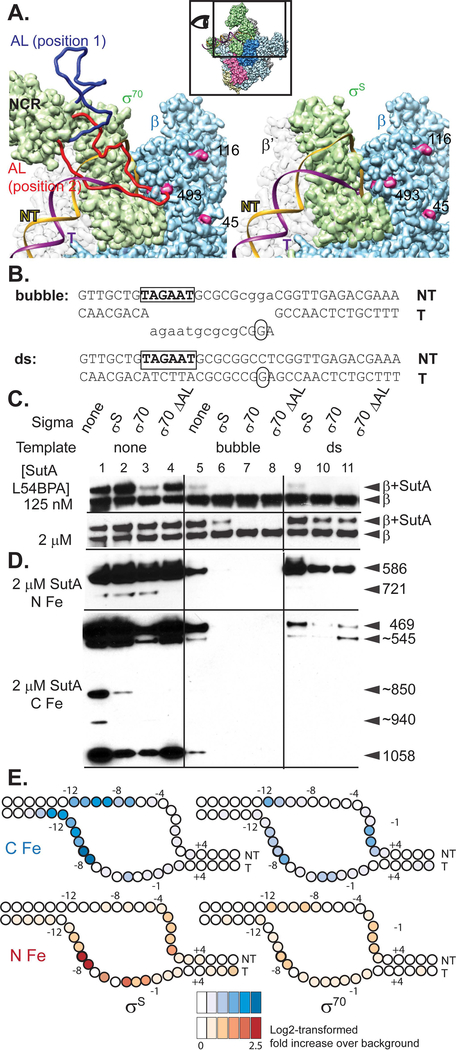
Figure 4. Both σ factor and DNA compete with SutA for binding to RNAP.
A. Models based on E. coli σ70 and σS holoenzyme structures. The inset shows the perspective and extent of this view relative to the holoenzyme structure shown in (2B). The P. aeruginosa β sequence was threaded onto an E. coli crystal structure (PBD: 5UAG), and then the β subunit from this was docked into the Eσ70 cryoEM structure (left; PDB: 6CA0) or the EσS crystal structure (right; PDB:5IPN). Residues showing cross-link or cleavage reactivity with SutA (Fig. 2) that are visible in this view are colored magenta and numbered. Residues 168–212 of σ70, which are not visualized in the cryoEM structure, were modelled in as a flexible loop (AL). Two possible loop positions are shown (red and dark blue), one of which (red) could clash with both the DNA and SutA positions. In contrast, σS does not appear likely to directly contact SutA (right). B. Sequence and structure of template DNA surrounding transcription start site. C. Western blot showing cross-linking of L54BPA SutA to β, in the context of different σ factors and promoter DNA. D. Western blots showing β cleavage mediated by N-Fe or C-Fe SutA FeBABE conjugates. Sizes of cleavage products were estimated by comparison to β fragments of known sizes analyzed on large non-gradient gels (Supporting Information, Fig. S12 and S21); for some products (~), only approximate sizes can be determined. The blot for C-Fe was exposed for longer (4 minutes) than the blot for N-Fe (30 seconds). E. Cleavage of the DNA in the rrn promoter complexes formed by Eσ70 or EσS in the presence of N-Fe or C-Fe SutA, revealed by primer extension. Average log2-transformed enrichment in signal between the FeBABE reaction and a negative control reaction containing unmodified SutA, from triplicate measurements, is represented by color intensity for each base. FeBABE reactions and the negative control reactions to which they were normalized were run on the same gel. The DNA is depicted as an almost completely opened transcription bubble, because in this conformation the strongest DNA cleavages for the N- and C-tail FeBABE SutA would be in close proximity to the strongest protein cleavages for these reagents. However, the cross-linking data suggest that SutA may be displaced by the fully stabilized open complex.