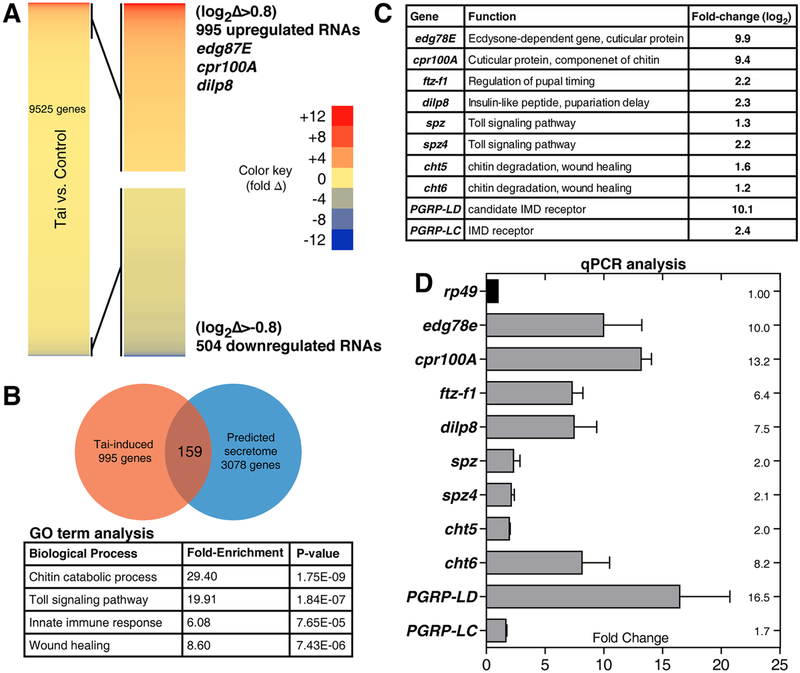
Figure 3. Identification of Tai-regulated mRNAs in larval and pupal wing discs.
(A) Heat-map of abundance changes in 9525 mRNAs in en>tai,GFP vs. en>GFP (Tai:Ctrl) L3 wing discs, with 995 upregulated >+0.8(log2)-fold and 504 downregulated <−0.8(log2)-fold. (B) Top GO-terms among 995 upregulated transcripts, with fold-enrichment and p-values. Venn diagram depicts 159 factors in overlap of Tai-induced mRNAs and predicted secretome (ref.[23]). (C) Fold-change of selected factors by RNA-HTS and (D) by qPCR of 6hr APF MS1096>tai wing disc RNAs standardized to rp49. Error bars = standard deviation (SD) of three biological replicates. See also Figure S2, Data S1 and Tables S1–4.