Figure 1.
Four-Way Host-Microbe-Drug-Nutrient Screens Identify a Signaling Hub for the Integration of Drug and Nutrient Signals
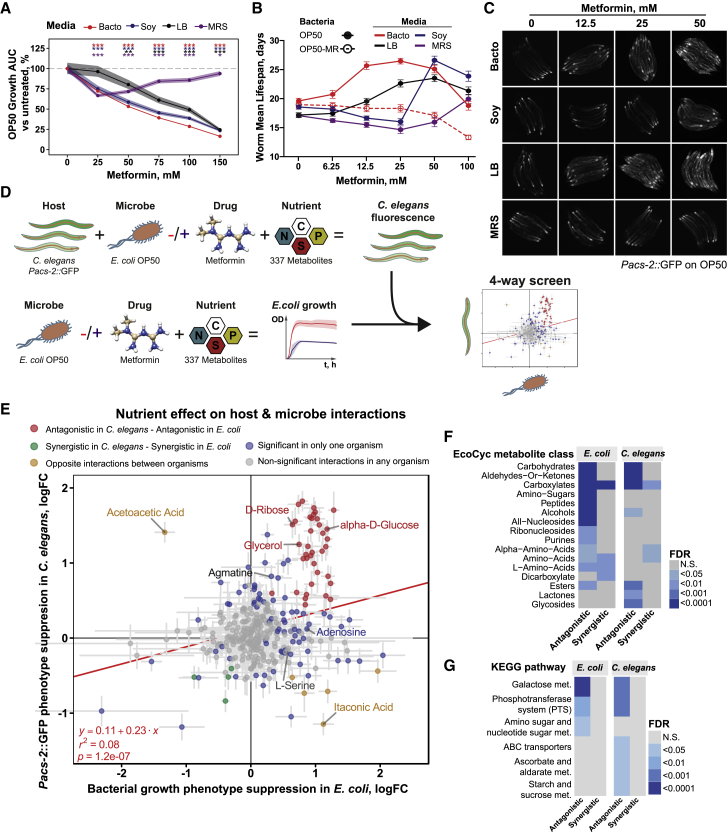
(A–C) The effects of metformin on bacterial growth (A), wild-type N2 worm lifespan (B), and metabolism (C) are dependent on drug dose, nutrients, and bacteria. OP50-MR is an E. coli OP50 strain that developed metformin resistance. As observed previously (Cabreiro et al., 2013), metformin does not extend the lifespan when worms are grown on OP50-MR. In (B), each data point corresponds to the mean lifespan of 80–154 worms. See also Table S1. In (C), each panel shows 8 individual worms.
(D) Diagram of the four-way host-microbe-drug-nutrient interaction screen.
(E) Nutrient effects on bacterial phenotype (growth, x axis) and on wild-type N2 worm phenotype rescue (Pacs-2::GFP expression, y axis) in response to metformin. The red fit line shows the correlation between metformin and nutrient effects in bacteria and worms. Antagonistic or synergistic refers to the type of interaction determined by linear modeling observed between metformin and nutrient effects, leading to an overall effect that is significantly greater than the sum of the effects of the two components alone either in C. elegans Pacs-2::GFP levels or E. coli growth. Positive fold changes indicate nutrient suppression of the effect of metformin in bacterial growth or C. elegans Pacs-2::GFP expression. Error bars represent SE. FDR < 0.05 for significance. All colored circles are statistically significant. Gray circles are non-significant. Effects of highlighted nutrients are provided in detail in Figures S1 and S2.
(F and G) EcoCyc metabolite class (F) and KEGG pathway (G) enrichment for the effects of nutrients on E. coli OP50 growth and worm Pacs-2::GFP expression in the context of metformin treatment.
Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Table S1 for lifespan statistics and Table S2 for screen statistics.