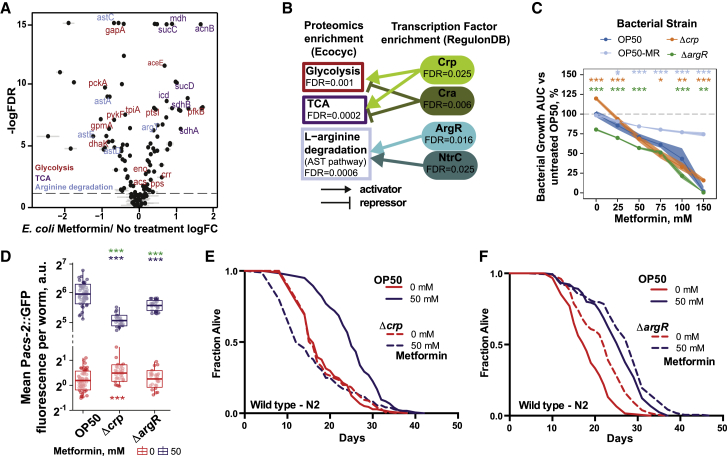
Figure 2.
Bacterial Proteomics Identify Transcriptional Networks Underlying Metformin Effects in E. coli
(A) Volcano plot showing E. coli proteins that are differentially regulated in response to metformin. Highlighted proteins belong to significantly enriched KEGG pathways.
(B) Diagram displaying connectivity between KEGG pathway enrichment and RegulonDB transcription factor (TF) enrichment from proteomics data of E. coli OP50 treated with metformin.
(C) Bacterial growth summary of E. coli OP50 TF mutants with metformin. Significance stars represent comparison with OP50 for each metformin concentration.
(D) Metformin regulates worm Pacs-2::GFP expression in a bacterial TF-dependent manner. Significance stars represent comparison with OP50 at 0 mM (red) or 50 mM (purple) and metformin-genotype interaction (green).
(E and F) Metformin extends worm lifespan in a bacterial TF-dependent manner.
Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Table S1 for lifespan statistics and Table S3 for proteomics statistics.