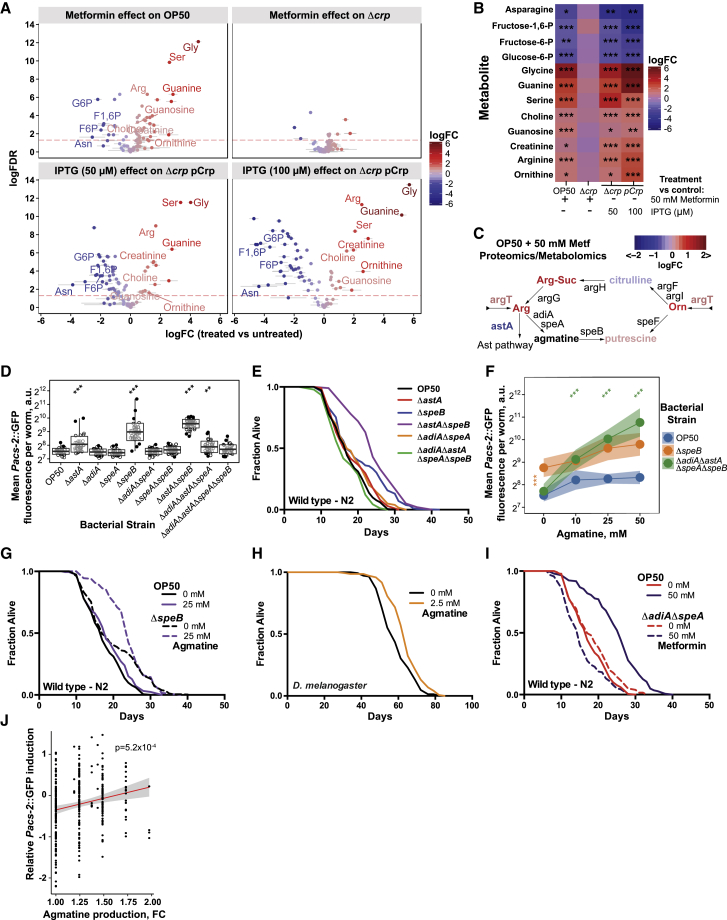
Figure 4.
Bacterial Agmatine Regulates Host Metabolism and Lifespan
(A) Volcano plots of metabolomics data showing effect of metformin in control E. coli OP50 or an OP50 Δcrp mutant and the effect of Crp overexpression in OP50.
(B) Subset of differentially and significantly expressed metabolites that are unique to Crp regulation and metformin treatment.
(C) Bacterial arginine-related metabolic pathways with an overlay of metformin-induced changes in the E. coli proteome and metabolome. Ast, arginine N-succinyltransferase pathway.
(D and E) Deletion of genes from E. coli arginine catabolism alters worm Pacs-2::GFP expression (D) and lifespan (E).
(F and G) Agmatine supplementation upregulates worm Pacs-2::GFP expression (F) and extends the lifespan (G) in a bacterium-dependent manner.
(H) Agmatine supplementation extends the fly lifespan in sugar-yeast-agar (SYA) medium.
(I) Metformin does not extend the lifespan in the agmatine-deficient OP50 mutant ΔadiAΔspeA.
(J) Comparison of in silico predicted agmatine production capacity and measured worm Pacs-2::GFP expression with nutrient supplementation in the context of metformin. The p values indicate the significance of association between predicted agmatine production capacity and Pacs-2::GFP fluorescence (linear model fit). See Figure S5J for predicted agmatine production capacity and measured growth-rescue of metformin-treated E. coli OP50.
Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Table S1 for lifespan statistics, Table S3 for proteomics statistics, and Table S4 for metabolomics statistics.