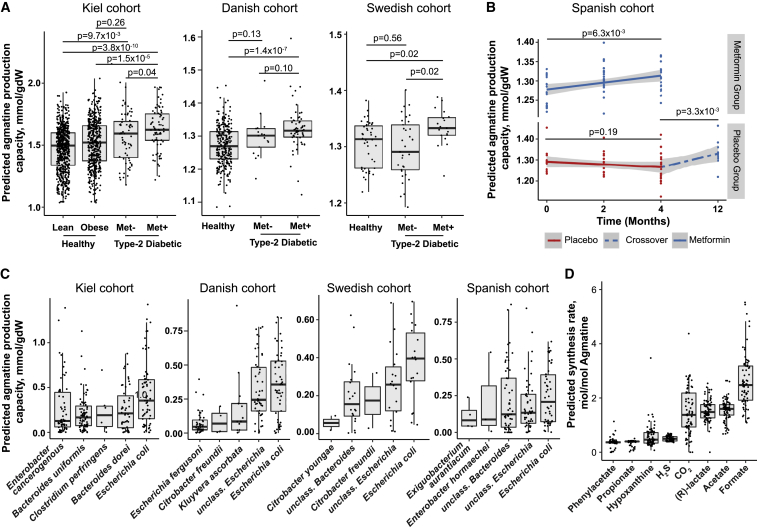
Figure 5.
Metabolic Modeling of Human Gut Microbiota Reveals Signatures of Agmatine Overproduction in Metformin-Treated Type 2 Diabetic Patients
(A) Predicted agmatine production by the gut microbiota in the 3 independent cohorts. Shown are FDR-corrected p values from Wilcoxon rank-sum tests between the indicated groups.
(B) Longitudinal changes in predicted agmatine production following initiation of metformin treatment in newly diagnosed type 2 diabetic patients. The p values indicate the significance of the treatment effect (i.e., time) on agmatine production (linear model fit).
(C) Predicted top 5 microbial producers of agmatine within the gut microbiome of metformin-treated patients across cohorts.
(D) Side products of predicted agmatine production in the Kiel cohort. Values correspond to moles of side product produced per mole of agmatine produced.
Data are represented as absolute values.
For details regarding statistical tests, see STAR Methods and Table S5. mmol/gM/day, predicted production fluxes in millimoles per gram of gut microbiota per day.