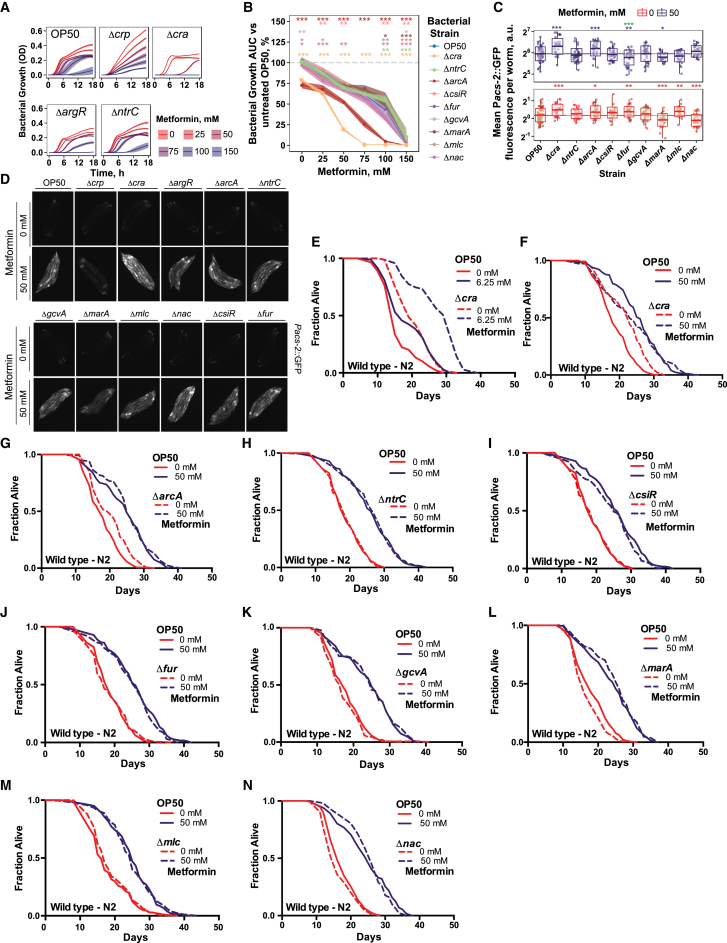
Figure S3.
Bacterial Proteomics Identify Transcriptional Networks Underlying Metformin Effects in E. coli, Related to Figure 2
(A) Bacterial growth curves of E. coli OP50 transcription factor (TF) mutants with increasing concentrations of metformin. Shaded area shows mean growth OD ± SD.
(B) Bacterial growth summaries of E. coli OP50 deletion mutants for TFs associated with proteomic changes in response to metformin treatment. Significance stars represent comparison with OP50 for each metformin concentration. Opposite to the effects of metformin on the resistant OP50-MR strain compared to OP50, Δcra and ΔarcA mutants exhibited increased sensitivity to bacterial growth inhibition by metformin.
(C and D) Metformin regulates worm Pacs-2::GFP expression in a E. coli OP50 TF-dependent manner. Worms grown on Δcra (A) and ΔarcA mutants (B) showed an increased activation of host Pacs-2::GFP expression in an additive manner to metformin. For C, significance stars represent comparison with OP50 at 0 mM (red) or 50 mM (purple) and metformin-genotype interaction (green). In (D), each panel shows 5 individual worms.
(E and F) Worm lifespan extension by metformin is enhanced with a Δcra E. coli OP50 mutant at low (6. 25 mM) (E) but not high (50 mM) (F) drug concentrations. As previously reported (Cabreiro et al., 2013), these data suggest a shift in the window of action of metformin on host longevity depending on the sensitivity of the bacterial strain to growth inhibition by metformin.
(G-N) Survival curves of E. coli OP50 TF mutants that do not affect worm lifespan extension by metformin.
Data are represented as mean ± SEM unless otherwise stated. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. See also Table S1 for lifespan statistics and Table S3 for proteomics statistics.