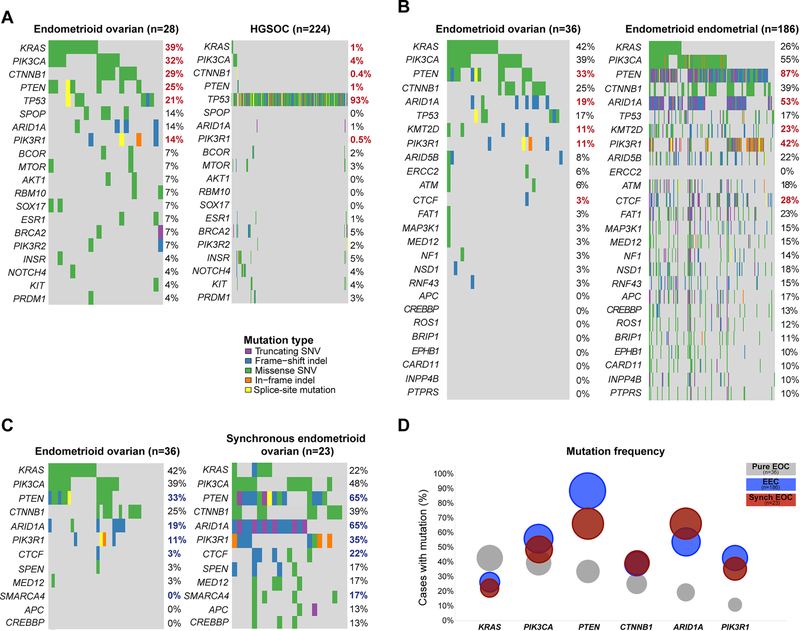
Figure 3. Comparison of the mutational profiles of pure endometrioid ovarian cancers with high-grade serous ovarian cancers, endometrioid endometrial cancers and synchronous endometrioid ovarian cancers.
The repertoire of somatic mutations affecting the 341 genes included in the MSK-IMPACT assay (smallest gene panel) in (A) pure microsatellite-stable/ POLE wild-type endometrioid epithelial ovarian cancers (EOCs) (n=28) and in high-grade serous ovarian cancers from The Cancer Genome Atlas (TCGA; n=224)[9] (B) all pure EOCs (n=36) and endometrioid endometrial cancers (EECs; TCGA; n=186)[8], and (C) all pure EOCs (n=36) and EOCs synchronously diagnosed with EECs (n=23)[14]. (D) Mutation frequencies of KRAS, PIK3CA, PTEN, CTNNB1, ARID1A and PIK3R1 in the pure EOCs studied here (n=36), in EECs (n=186, TCGA)[8] and EOCs synchronously diagnosed with EECs (synchronous EOCs; n=23)[14]. Comparisons were performed using Fisher’s exact test. (A and B) Benjamini-Hochberg adjusted statistically significant p-values (adjusted p<0.05) are shown in red bold font, (C) unadjusted statistically significant p-values (p<0.05) are shown in blue bold font.