Figure 4.
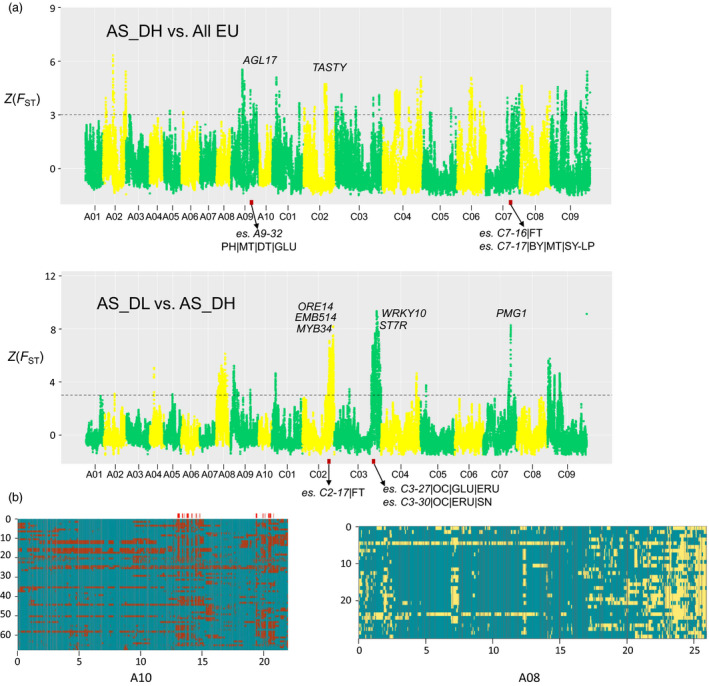
Genomic selection signals in Asian rapeseeds. (a) Significantly divergent genomic regions of Asian double‐high rapeseeds (AS_DH) relative to the European rapeseeds (EU) are shown on the top of the subfigure, and significant divergent genomic regions between Asian double‐low rapeseeds (AS_DL) and AS_DH are shown at the bottom. Selected genes and QTL found previously in those regions are indicated. (b) Local ancestry inference for Asian rapeseed. Left: Local ancestry inference for the 68 Asian rapeseeds in chromosome A10 (red: B. rapa genetic background; blue: EU genetic background). The red bars on the top indicate that the introgression events from B. rapa into the Asian rapeseed population have been almost fixed (i.e. found in >90% samples). Right: Local ancestry inference for the 30 Asian double‐low rapeseeds in chromosome A08 (blue: EU genetic background; yellow: Asian double‐high genetic background). The X‐axis represents the genomic locations of each chromosome. The Y‐axis presents representative individuals used for the ancestry inference. For details of all chromosomes, see Figure S14.
