Abstract
In the last decades, the amount of medical imaging studies and associated metadata has been rapidly increasing. Despite being mostly used for supporting medical diagnosis and treatment, many recent initiatives claim the use of medical imaging studies in clinical research scenarios but also to improve the business practices of medical institutions. However, the continuous production of medical imaging studies coupled with the tremendous amount of associated data, makes the real-time analysis of medical imaging repositories difficult using conventional tools and methodologies. Those archives contain not only the image data itself but also a wide range of valuable metadata describing all the stakeholders involved in the examination. The exploration of such technologies will increase the efficiency and quality of medical practice. In major centers, it represents a big data scenario where Business Intelligence (BI) and Data Analytics (DA) are rare and implemented through data warehousing approaches. This article proposes an Extract, Transform, Load (ETL) framework for medical imaging repositories able to feed, in real-time, a developed BI (Business Intelligence) application. The solution was designed to provide the necessary environment for leading research on top of live institutional repositories without requesting the creation of a data warehouse. It features an extensible dashboard with customizable charts and reports, with an intuitive web-based interface that empowers the usage of novel data mining techniques, namely, a variety of data cleansing tools, filters, and clustering functions. Therefore, the user is not required to master the programming skills commonly needed for data analysts and scientists, such as Python and R.
Keywords: PACS, Business Intelligence, DICOM, Data Analytics, Cloud, Big data
Introduction
Nowadays, medical imaging repositories contain a wide range of valuable metadata that describes thoroughly all the stakeholders involved in medical imaging practice. Despite being mostly used for supporting medical diagnosis and treatment, many recent initiatives claim the utility of medical imaging studies in clinical research scenarios and in the improvement of the medical institutional business practices.
The current paradigm of medical imaging repositories fits well with the definition of big data [1]. The continuous production of huge volumes of data, its heterogeneous nature, and the increasing number of performed examinations make the analysis of medical imaging repositories very difficult for conventional tools. Moreover, the new trend of distributed Picture Archive and Communications Systems (PACS) architectures that makes possible to federate multiple institutions in the same PACS archive at cloud [2] promotes the creation of large and more useful datasets. Therefore, DA and BI techniques applied to this scenario have potential to increase the efficiency and quality of the medical practice.
This article proposes an ETL framework for medical imaging repositories that feeds, in real-time, a BI platform oriented to medical imaging practice and research. The solution can index distinct data sources and aims to provide the necessary environment for conducting research on top of live institutional repositories. It leverages all the metadata stored in those repositories without requiring a data warehouse, predefined data models, or imposing rigid data flows. The developed system takes advantage of Dicoogle’s data mining features [3] for extracting data from production PACS and provides a series of exploratory techniques and visualization tools for a deep understanding of the working dataset and extraction of valuable information. Moreover, its design facilitates the use of analytics tools without requiring user programming skills commonly used in other platforms (e.g., Python and R). It provides an intuitive Web-based interface that empowers the usage of novel data mining techniques, namely, a variety of data cleansing tools, filters, and clustering functions. Moreover, it features an extensible dashboard with customizable charts and reports.
Background
DICOM
The proliferation of PACS was possible mostly due to the development of Digital Imaging and Communications in Medicine (DICOM), the standard for handling of medical imaging data. PACS are responsible for providing storage, transmission, and even printing services, among others, to allow connectivity, compatibility, and workflow optimizations between different medical imaging equipment [4].
The DICOM standard supports not only the pixel data that defines the medical images but also a wide range of metadata information related to all the stakeholders involved in the clinical practice, such as the patient, procedure, equipment, staff-related data, or structured report. Data relative to these stakeholders is conveyed by DICOM data elements which compose DICOM objects or files.
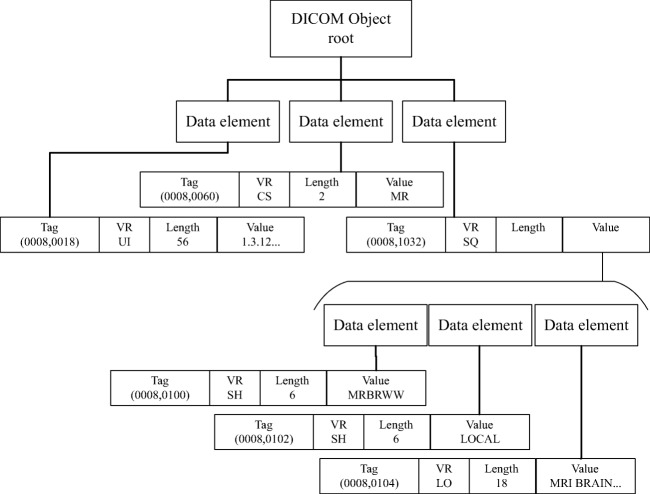
DICOM data elements are encoded using a Tag-Length-Value (TLV) structure. The tag field identifies the data element and includes two subfields: the group identifier and the element identifier within the group, both encoded using 16-bit unsigned values. DICOM data elements are grouped by their relation with real-world entities, i.e., Information Entities (IE) that represent, for instance, the patient (0 × 0010), the study (0 × 0008), and the series (0 × 0020). Therefore, elements holding information related to the patient are encompassed in the patient group (0 × 0010) and so on. As an example, the patient’s name tag is represented by (0 × 0010, 0 × 0010). Apart from the tag, DICOM data elements include also the field length in bytes. Lastly, the value field holds the actual element’s data. A simple illustration of a DICOM data element is presented in Fig. 1.
Fig. 1.
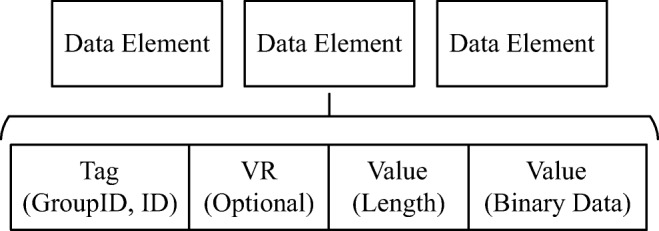
DICOM data elements structure
DICOM object is an umbrella term to describe a DICOM file, which could be images, structure reports, among others. The information enclosed in DICOM objects is very heterogeneous. There are data elements for representing names, measures, dates, among others. Therefore, to express all these data types, the encoding of the value field changes according to the element’s type. The part 5 of DICOM standard [5] defines 27 different encoding formats for the value field. These are the most basic data types in the standard and are called Value Representation (VR). A data element can include sub-group of elements, having the SQ (Sequence) VR. This creates a hierarchical document structure similar to many contemporary data models. This structure is illustrated in Fig. 2. The data element’s VR can be declared explicitly by inserting a VR field into the element’s TLV structure, thus turning it into Tag-VR-Length-Value, or alternatively, it can be implicitly inferred by reading the DICOM standard dictionary [6] that contains near of 2000 entries [7]. These elements cover very well the general requirements of medical imaging environments. Nonetheless, the standard is extensible and private data elements may be added by manufacturers to support their latest features. Thus, private dictionaries may extend the default provided by the standard. By doing so, DICOM standard has the capability of keeping up with state of the art solutions, a fundamental aspect for its prevalence in the field.
Fig. 2.
Illustration of nested objects in DICOM
DICOM objects can represent multiple medical imaging artifacts, such as images from a wide range of modalities or structured reports (SRs). These objects are composed of several modules associated with an IE, which represents real word stakeholders such as patients and studies. They include multiple data elements; for instance, the patient module is composed of the patient’s name, sex, birthday, among other attributes.
DICOM also defines which modules shall be included for each DICOM object class, as per its specific information model definition (IOD) [7]. IOD are collections of modules that describe each object. They define which information must be included in the DICOM file, along with its type.
DICOM includes the concept of instances that are IOD templates filled with real-world data and are identified by unique identifiers (UIDs). Every DICOM object’s instance UID must be unique.
Finally, the standard provides a DICOM Information Model (DIM) that follows the hierarchical organization in patient-study-series-image. This approach resembles the real-world organization since a patient may perform multiple studies, each study may have several series of many modalities and each procedure may result in a large collection of image instances. Since DICOM images are typically spread by multiple files for convenience. The linkage between these entities is achieved using the instances UID. Consequently, the DIM may be reconstructed even when images are spread across multiple repositories. The UID attribute is generally named after its Stakeholders name; however, there are exceptions like the SOPInstanceUID (ServiceObjectPair) which is the UID of the whole DICOM object.
Imaging Business Intelligence
The analysis of the PACS archives content has been demonstrated to withdraw positive outcomes for many research endeavors, namely, radiation dosage surveillance [8], performance analysis of institutional business practices processes [9, 10], the cost-effectiveness of diagnostic procedures [11], among many others [11–14].
Nevertheless, the complexity of medical imaging data has increased tremendously [15] since the volume and heterogeneity of data generated by the medical practice also increased considerably. Therefore, researchers must rely on Information Technologies (IT) tools to be able to perform their analytical tasks. However, traditional PACS do not allow exploring the imaging metadata for extraction of relevant knowledge. This leads to the use of third-party applications to perform data analysis, including proprietary solutions that only work with specific PACS.
BI consists of a pipeline that integrates a series of tasks. When combined, these are responsible for acquiring, transforming, and translating raw data into useful information for improving the business practices [16]. This process encapsulates a multitude of capabilities such as reporting, dashboards, and data mining [17].
Nowadays, the data generated by medical imaging laboratories is highly heterogeneous and inconsistent. Although all equipment implements the same DICOM standard, they might use different configurations. This generates irregular data, which occurs, for instance, when two different equipment report the same value but using different metrics [18]. Furthermore, the interaction between technical staff and equipment is another inconsistency factor. For these reasons, applying analytical procedures to inconsistent data may generate unreliable results. So, one of the most important and crucial steps in the whole BI process is the Data Cleansing (DC) stage. It ensures the reliability of the data in each repository, by detecting and correcting inaccurate records [19]. DC is a complex process (Fig. 3), which can be further divided into a series of iterative operations. First, the Data Auditing step, where the working dataset is analyzed to determine which anomalies it contains. Next, the Workflow Specification step defines a set of operations required for fixing the previously identified anomalies (or, in some critical cases, exclude them). After this, the operations are executed in the Workflow Execution step. Finally, the applied operations are validated in the Post-Processing and Controlling step. Each of these steps can be performed manually, supervised or unsupervised in line with the level of user input system required.
Fig. 3.
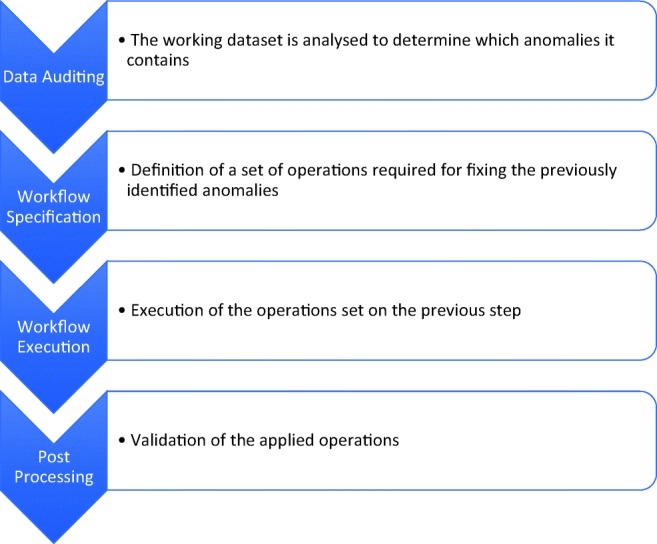
Data cleansing process
Dicoogle
Dicoogle is an open source PACS archive that has an extensible indexing and retrieval document-based system [20]. It provides easy expansion of new functionalities through the development and usage of plug-ins using the available Software Development Kit (SDK). The disposal of the SDK allows developers to expand the functionalities without making changes in the core of the system.
Dicoogle was designed to support information extraction, indexing, and storage of the metadata contained in DICOM files without any special reconfiguration requirements [21].
This extendible architecture enabled the use of Dicoogle in research and healthcare industry and as an educational tool [2, 22] since the need to improve, monitor, and measure the medical imaging systems along with the extraction of knowledge from the medical images including healthcare quality indicators is of importance.
Furthermore, Dicoogle is being used as a support platform for DICOM data mining [22]. The DICOM data mining tools proven to be valuable to gather relevant data for the professional healthcare improvement by identifying factors that may contribute to a quality deficit [23].
Related Work
There are some references in the literature relative to the usage of DA and BI systems applied to medical imaging repositories. Nagy et al. [12] developed a tool that implements a fully fledged BI stack. It starts by aggregating data from the institution’s systems, namely, DICOM metadata from the PACS and Health Level 7 (HL7) data from the Radiology Information System (RIS), among others. Data is extracted on a periodical basis and stored in a MySQL database that feeds a dashboard. It is a graphical tool that includes the most relevant chart types, such as histograms and bubble charts. Whenever a database’s entry is added or updated, the corresponding chart is automatically rendered to reflect these changes, as well as the details of currently selected reports.
Kallman et al. [20] developed a framework that provides a similar set of functionalities. However, there are some important differences. For instance, the second framework separates the DICOM metadata header from the image’s pixel data, storing the first in a separate repository. Moreover, the user interface is command line based on Structured Query Language (SQL). It is a powerful and flexible language for database experts but harder to perform data analysis for less experienced users, such as radiology researchers.
The two previous frameworks make use of separate repositories; i.e., they do not work directly over institutional PACS archives. The advantage is that they can be used for statistical purposes with no risk of interfering with the regular clinical workflow. However, they will always work with an outdated snapshot of the archive’s content. In our point of view, the major limitation is that they are bound to a strictly relational database model. This means that researchers cannot derive knowledge from metadata fields that were not previously encompassed in the database schema, not satisfying this way the requirements of most research endeavors.
Wang et al. [8] developed a database solution for controlling patient’s radiation exposure by analyzing the DICOM images metadata. It is focused on the detection of irregular radiation dosages by several filters and can aggregate information at the study, patient, and institutional level. It includes reporting, alerts capabilities, and a Web interface for interacting with the system. The most interesting module, in the context of this article and comparing with the previous tools, is the Knowledge Base that is capable of unifying distinct vendor data by enforcing the measured attributes to use the same units, which were defined statically. However, the major constraint of this solution is that it cannot be used in other imaging contexts.
In [18], the authors present a DICOM Data Warehouse for arbitrary data mining. It enables automated data analytics tasks on top of DICOM metadata databases. The authors claim that despite existing some previous efforts in the literature, namely [8, 9], none shared the purpose of enabling completely arbitrary data mining (DM) capabilities. The solution is backed by a relational database, which data model needs to be extended to support new Data Elements. It also targets the discrepancies between data attribute’s values from different vendors by creating static mappings for different attributes that represent the same measurements. However, this framework does not provide a graphical interface for creating reports, alerts, or browsing the data. In an updated article [21], the authors point out the difficulty of indexing a very heterogeneous data source, such as DICOM, which resulted in leaving some DICOM attributes unindexed. They state that Not only SQL (NoSQL) approaches may be required to handle it.
Architecture
The proposed architecture follows a classic client-server model, segmented into three distinct layers: presentation, business, and persistence. In this architectural pattern, the client acts only as the presentation layer of the developed system, displaying the interface to the user and resorting to the server when required. In turn, the server is responsible for implementing the business and persistence layers of the application. The business layer encompasses most of the application’s logic, handling the client’s requests and providing an adequate response. The persistence layer is responsible for storing and maintaining data across multiple sessions.
The server’s BI functionalities are provided by the Python Scipy stack (which includes the NumPy, SciPy, and pandas libraries) and the scikit-learn library. The methods developed using the previous libraries are exposed through a Representational State Transfer (REST) application interface (API), using Django and the Django REST framework toolkit.1 The persistence layer is based on PostgreSQL Relational Database Management System (RDBMS), which persists the application’s logic-related data, in conjunction with panda’s Hierarchical Data Format Store (HDFS), responsible for bulk data storage.
The client is a Web application that follows the single page application pattern, in which all the necessary client code (HyperText Markup Language (HTML), JavaScript (JS), and Cascading Style Sheets (CSS)) is loaded in a single page load. The application was developed using the React framework, with the help of the Redux state container. Furthermore, it makes use of the Bootstrap framework to manage most of the interface as well as the plotly.js library to handle charts rendering.
This framework leverages the Dicoogle’s data mining features for extracting data from the DICOM objects in the PACS. The communication is supported by the Dicoogle’s content discovery and retrieval services, namely, its Web service query end-points. In turn, these services rely on the technologies implemented by plug-ins, as described in [22].
Despite the development of the dashboard, the presented architecture, through the business and persistence layers, allows the supplying of data to third party framework (e.g., Kibana2 or Grafana3).
Figure 4 presents the functional modules that will be described in the next section, as well as their interactions.
Fig. 4.
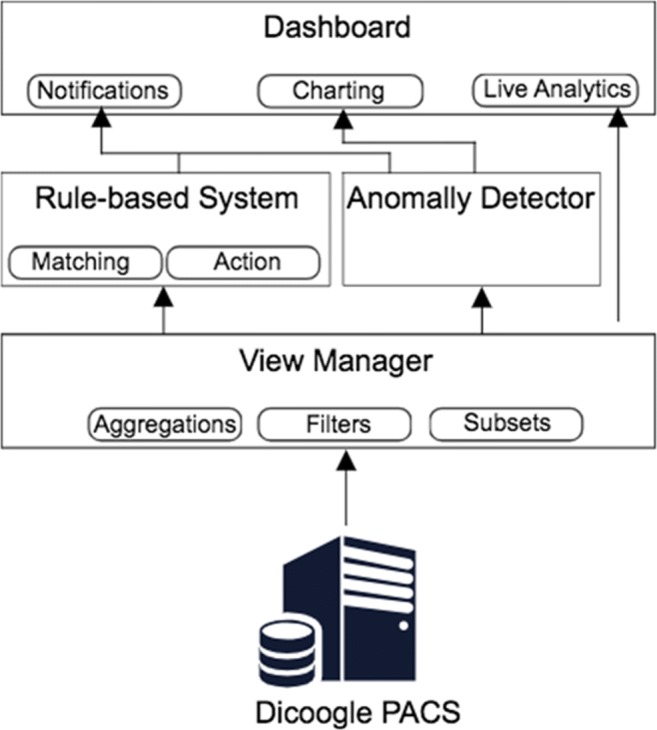
Business Intelligence framework architecture
Rule-Based Data Cleansing
One major concern was to allow the manipulation of multiple irregular records simultaneously because an inaccuracy occurs multiple times in the same dataset. Consequently, manual correction of those records would be impractical. To avoid that, a Rule-based control system was devised for supporting Data Cleansing features. It works in two phases: Matching and Action. In the first phase, the different rules are applied in the dataset to detect inaccurate records. The action phase performs the corrections to the records.
A set of basic rules and actions were developed to provide a powerful service and keeping, at the same time, the usability high. The rules cover most use cases defined in the context of this work but it can be extended to cover others by adding additional modules, written in Python, which are interpreted at runtime.
The following five rules are currently available in the system:
Empty Field: allows the detection of empty values on a set of fields. This issue is the most common anomaly when working with data. Unfortunately, most of these fields cannot be inferred, usually leading to their removal from the dataset;
Expected Value: for detecting one (or more) values on a given group of fields. It works by defining a set of values and the fields where those values are expected;
Regular Expression: a more generic and advanced version of the Expected Value rule, requiring Regular Expression’s knowledge;
Filter Date: selects a set of records on which the Date field matches the defined interval;
Expression: enables the execution of statements or even small scripts, allowing to detect more complex cases that are impossible to perform with the previous rules. It should only be used as a fallback.
Previous rules can be combined with the following actions:
Fill Empty Field: it replaces the empty values in the defined fields with a chosen value;
Replace by Value: it replaces a defined set of values in a given range of fields;
Replace by Regex: a specific case of the previous action when the value, to be replaced, is a Regular Expression;
Date to Age: it converts a specified field (usually the Patient Date of Birth field) to the corresponding Age (most often the Patient Age field) according to the DICOM standard;
Normalize Age: convenience action to normalize the Age field, enforcing the measure to be in Years, Months, Weeks or Days;
Expression: it accepts a python script to implement a free action.
It is important to note that defined rules might not be mutually exclusive since a rule’s triggered action might update the values that match another rule. For this reason, every rule has a priority attribute, an integer value, and the rules are executed according to their priority.
Anomaly Detector
This component permits to detect anomalies in data by associating a given field to one of two categories: ordinal or nominal data, based on the field’s datatype. As the names imply, if the selected field’s datatype is numerical (either integer or float), then it is provided with an ordinal description; otherwise, it is provided with a nominal description.
An ordinal description returns a count attribute, with the total number of non-empty entries in the field, as well as some of the most common statistics for a numerical set of values, such as mean, standard deviation, minimum and maximum values, and percentiles (25%, 50%, and 75%). On the other hand, a nominal description returns the number of entries in which a given value appears. Also, both descriptions feature an empty values’ count, if applied.
Unfortunately, the automatic detection of a field’s description based on its datatype is sometimes flawed. Often, a field is composed of an integer set of values. However, those values do not represent a continuous variable, meaning that they are supposed to be interpreted as a category. Therefore, it is possible to manually enforce a nominal description for fields composed entirely of integer values.
View Manager
The concept of Views was implemented to ease the process of working with large and heterogeneous datasets. Most use cases do not work with all repository data, particularly, when the size and heterogeneity of data would affect significantly the performance of most operations, even the most basic ones. Moreover, very often the analysis targets a specific subset of the repository, such as analyzing data related to a specific modality or analyzing reports performed in a given date range. Taking this into consideration, it was developed a view concept that intends to represent a smaller and more specific dataset from the original PACS repository.
Therefore, Views are very important because they determine the execution context of the other components, i.e., rules and anomaly detectors will only be applied to their assigned Views, and consequently sub-datasets. It allows the user-created Views to inherit the transformations of their parent views, much like the concept of Object-Oriented (OO) inheritance. User-created Views can inherit either from the default (root) View or from other user-created View. This provides flexibility and extensibility to the system. The Views support all previously referred data manipulations. For supporting this, the system allows merging a view’s transformations with its parent’s, thus optimizing the usage of the Dicoogle’s Data Mining capabilities.
To create new views, three transformations are provided:
Aggregation: it mimics the SQL group by operation, defining the set of aggregation fields and functions to be performed. It is possible to provide only one function as a parameter. In this case, it will be applied to all the fields that are not part of the aggregation fields. However, this is rarely the desired output. For this reason, it is also possible to define one or more functions per column. This possibility is particularly useful for taking into consideration the Information Entities Hierarchy (Patient, Study, Series, Image). The allowed functions are cumulative sum, cumulative product, maximum, minimum, median, standard deviation, mean, size and sum;
Subset: it enables the creation of a static subset, defining the interval of either rows and/or fields (i.e., selection and projection restrictions);
Filter: it makes possible to use the above rules, as transformations.
The defined transformations can be applied using the Dicoogle data mining capabilities in two situations. Firstly, when an Expected Value filter is specified, it is translated exactly to the Dicoogle’s query language. For instance, a filter by Modality and PatientSex, and expected values CR and F respectively, would be translated to “Modality:CR AND PatientSex:F” in the Dicoogle’s query service.
Secondly, when the fields of a subset transformation are defined they are also mapped to the Dicoogle’s query interface. For instance, the subset with fields Modality and PatientAge, are translated to the query’s return fields = [Modality,PatientAge].
Dashboard
The developed solution provides extensive dashboard capabilities. The dashboard enables the development of a fully customizable client page that may include any of the available visualization components, each one inserted in a fully resizable and sortable panel. Visualizations also depend on the defined views. So, the system not only stores the layout of the dashboard, including every panel’s coordinates and size, but also the instructions necessary to populate the component with the desired data.
Given its integration with the underlying Dicoogle PACS, the proposed system provides real-time analytics capabilities. This means that the platform is immediately notified when new data arrives at the archive, avoiding the necessity of creating repository snapshots, acquired on a periodical basis. New images are automatically added to the necessary Views and are properly analyzed by the previously defined rules. Later, a notification is also sent to the Dashboard with updated information.
Results
This section demonstrates the content discovery capabilities of developed framework for extracting knowledge from medical imaging repositories. This demonstration was based on an affiliated institution PACS archive with roughly 35 million files. Dicoogle BI was deployed and used in parallel with institutional PACS. In the scope of this work, sensitive data was anonymized using an in-house tool so that it would not be disclosed.
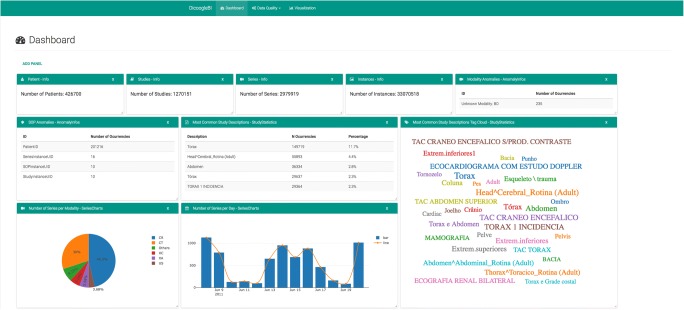
An overview of the Dicoogle BI dashboard is shown in Fig. 5, including several widgets. On the top, there are four informative widgets that count the number of patients (426700), studies (1270151), series (2979919), and instances (33070518) in the dataset. Initially, we noticed a discrepancy between these values and previous values generated by an earlier and simpler tool. This tool simply performed a count of the identifier attribute of images (35476975) and patients (279392).
Fig. 5.
Overview of the Dicoogle BI dashboard
The discrepancy between the number of images and the number of instances (identifier) was caused by duplicated DICOM files in the repository. This can be explained by the fact that some software platforms make copies of original files (for instance, produce images with lower resolution for fast display purpose) without having their SOPInstanceUID updated, which violates the standard, and caused the previously overestimation of the number of images in the repository. It also had an impact on the estimation of the number of the other stakeholders.
The dashboard includes also two widgets related to anomalies in the dataset. The first finds unknown Modalities, i.e., modalities that are not defined in the standard. For instance, it was identified 235 different series from a modality called OT (Other). The second widget identifies duplicated identifiers in the dataset. At the patient level, it was also identified discrepancies caused by duplicated identifiers for clearly different records. For instance, the same PatientID was given for images with different (PatientName, PatientBirthDate, PatientSex) tuples. In this case, over 201,216 cases where identified,4 representing 0.6% of repository instances. Although many proprietary PACS can work around these inconsistencies, they do not favor any analytical endeavor that relies on the quality of the collected data.
Other widgets with statistics were also presented in the Dashboard. For instance, there is a pie chart summarizing the contents of the dataset according to the series modality. There is also a bar chart with the production of series per day. Notice the reduced productivity on weekends (11th and 12th July, and 18th and 19th July) compared to regular weekdays. It is also perceivable the impact of the holidays on the institution productivity level, and an unexplainable decrement on Friday compared to the other weekdays.
Lastly, a tag cloud widget with study descriptions was included. It is possible to observe that the StudyDescription attribute has a lack of normalization. The previous statistics required the aggregation of the dataset by SeriesInstanceUID and then by Modality before the count function could be applied.
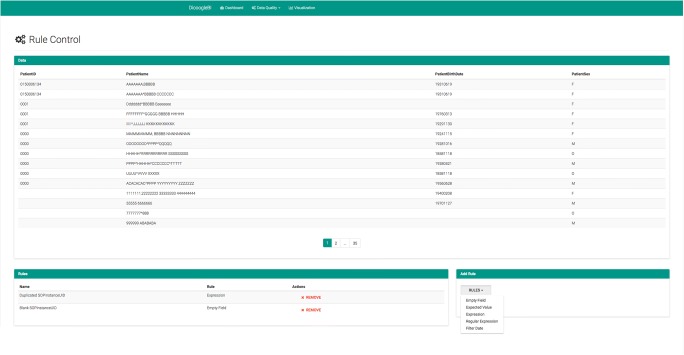
Figure 6 shows the interface for configuring the rules of a given view. The sample data shows the duplicated PatientID records referred in a previous paragraph. The records are obfuscated by privacy reasons, but a real name will be always replaced by the same obfuscated version. Thus, you can see clear different PatientNames with the same identifier. Very suspicious PatientBirthDates can also be seen, for example, 18581118.
Fig. 6.
Sample of rules applied to a view of the dataset
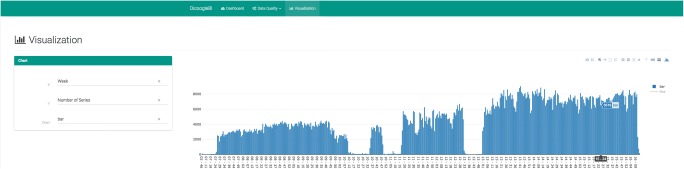
Finally, an overview of the application charting interface is shown in Fig. 7. The attributes and chart type can be customized for a given view. The chart itself is interactive, being possible to resize the visualization window and apply transformations to the axis. The presented graphical functionalities allow any user to perform DA and BI tasks on their PACS content even if they do not have programming skills.
Fig. 7.
Overview of the Dicoogle BI visualization interface
The capabilities of the Dicoogle’s BI module can also be applied to other third-party PACS thanks to the Dicoogle’s ability of indexing other PACS archives content. Moreover, it could be used to support these research endeavors in nonproduction PACS. Although, the benefit of having a real-time analytical pipeline would be lost in both these cases.
Conclusion
Nowadays, the exploitation of medical imaging laboratories has become a crucial part of healthcare institutions business models. Production medical imaging repositories hold a tremendous amount of data. Since this data is derived directly from the medical practice, it has extreme accuracy for analytics purposes. Timely exploitation of this data has promised to improve the efficiency of medical institutions business practices, as well as the quality of healthcare services. Even though this importance has already been acknowledged by the community, the volume and production rate of medical imaging data makes manual analysis impractical.
The Dicoogle Business Intelligence framework, described in this document, addresses this problem. It enables the development of automated analysis’ workflows performed directly on top of live institutional repositories without requesting the creation of dedicated data warehouse. Business managers and healthcare researchers can automatically derive knowledge over large repositories, which previously would take simply too much time. Moreover, it has the capability of improving the repository quality by using both its complete rule system that provides all the necessary Data Cleansing functionalities, as well as the versatile View control. These tools can be complemented by the provided data analysis components, such as charts and anomaly detection module. Finally, these components can be further combined on the application’s Dashboard, allowing the operator to further tailor his own workflow.
Funding Information
Tiago Marques Godinho is funded by Fundação para a Ciência e Tecnologia (FCT) under grant agreement SFRH/BD/104647/2014. Rui Lebre received support by the Integrated Programme of SR&TD “SOCA” (Ref.CENTRO-01-0145-FEDER-000010), co-funded by Centro 2020 program, Portugal 2020, European Union, through the European Regional Development Fund. This work has received support from the ERDF European Regional Development Fund through the Operational Programme for Competitiveness and Internationalization, COMPETE 2020 Programme, and by National Funds through the FCT, Fundação para a Ciência e a Tecnologia within the project PTDC/EEI-ESS/6815/2014.
Footnotes
The same PatientID was given for records with different PatientName, PatientBirthDate, PatientSex tuples
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tiago Marques Godinho, Email: tmgodinho@ua.pt.
Rui Lebre, Phone: (+351) 234 370 200, Email: ruilebre@ua.pt.
João Rafael Almeida, Email: joao.rafael.almeida@ua.pt.
Carlos Costa, Email: carlos.costa@ua.pt.
References
- 1.Hamilton B. Big data is the future of healthcare. Teaneck: Cognizant; 2012. [Google Scholar]
- 2.Godinho TM, Viana-Ferreira C, Bastiao Silva LA, Costa C. A routing mechanism for cloud outsourcing of medical imaging repositories. IEEE J Biomed Health Inform. 2016;20(1):367–375. doi: 10.1109/JBHI.2014.2361633. [DOI] [PubMed] [Google Scholar]
- 3.Santos M, Bastiao L, Costa C, Silva A, Rocha N. “Clinical Data Mining in Small Hospital PACS: Contributions for Radiology Department Improvement,” in Information Systems and Technologies for Enhancing Health and Social Care. Hershey: IGI Global; 2013. pp. 236–251. [Google Scholar]
- 4.Mildenberger P, Eichelberg M, Martin E. Introduction to the DICOM standard. Eur Radiol. 2002;12(4):920–927. doi: 10.1007/s003300101100. [DOI] [PubMed] [Google Scholar]
- 5.Digital Imaging and Communications in Medicine (DICOM) Part 3: Information object definitions, NEMA, Standard, 2017.
- 6.Digital Imaging and Communications in Medicine (DICOM) Part 7: Message Exchange, NEMA, Standard, 2017.
- 7.Pianykh OS. Digital Imaging and Communications in Medicine (DICOM): a practical introduction and survival guide. Berlin: Springer Science & Business Media; 2009. [Google Scholar]
- 8.Wang S, Pavlicek W, Roberts CC, Langer SG, Zhang M, Hu M, Morin RL, Schueler BA, Wellnitz CV, Wu T. An automated DICOM database capable of arbitrary data mining (including radiation dose indicators) for quality monitoring. J Digit Imaging. 2011;24(2):223–233. doi: 10.1007/s10278-010-9329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu M, Pavlicek W, Liu PT, Zhang M, Langer SG, Wang S, Place V, Miranda R, Wu TT. Informatics in radiology: efficiency metrics for imaging device productivity. RadioGraphics. 2011;31(2):603–616. doi: 10.1148/rg.312105714. [DOI] [PubMed] [Google Scholar]
- 10.Ondategui-Parra S, Erturk SM, Ros PR. Survey of the use of quality indicators in academic radiology departments. Am J Roentgenol. 2006;187(5):W451–W455. doi: 10.2214/AJR.05.1064. [DOI] [PubMed] [Google Scholar]
- 11.Raghupathi W, Raghupathi V. Big data analytics in healthcare: promise and potential. Health Inf Sci Syst. 2014;2(1):3. doi: 10.1186/2047-2501-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagy PG, Warnock MJ, Daly M, Toland C, Meenan CD, et al. Informatics in radiology: automated Web-based graphical dashboard for radiology operational business intelligence. RadioGraphics. 2009;29(7):1897–1906. doi: 10.1148/rg.297095701. [DOI] [PubMed] [Google Scholar]
- 13.Andreu-Perez J, Poon CCY, Merrifield RD, Wong STC, Yang G-Z. Big data for health. IEEE J Biomed Health Inform. 2015;19(4):1193–1208. doi: 10.1109/JBHI.2015.2450362. [DOI] [PubMed] [Google Scholar]
- 14.Viceconti M, Hunter P, Hose R. Big data, big knowledge: big data for personalized healthcare. IEEE J Biomed Health Inform. 2015;19(4):1209–1215. doi: 10.1109/JBHI.2015.2406883. [DOI] [PubMed] [Google Scholar]
- 15.Langer SG. Challenges for data storage in medical imaging research. J Digit Imaging. 2011;24(2):203–207. doi: 10.1007/s10278-010-9311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watson HJ, Wixom BH. The current state of business intelligence. Computer. 2007;40(9):96–99. doi: 10.1109/MC.2007.331. [DOI] [Google Scholar]
- 17.Chen H, Chiang RHL, Storey VC. Business intelligence and analytics: from big data to big impact. MIS Q. 2012;36(4):1165–1188. doi: 10.2307/41703503. [DOI] [Google Scholar]
- 18.Langer SG. A flexible database architecture for mining DICOM objects: the DICOM data warehouse. J Digit Imaging. 2012;25(2):206–212. doi: 10.1007/s10278-011-9434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang JTL, Zaki MJ, Toivonen HTT, Shasha D. Introduction to Data Mining in Bioinformatics. In: Data Mining in Bioinformatics. London: Springer; 2005. pp. 3–8. [Google Scholar]
- 20.Valente F, Silva LAB, Godinho TM, Costa C: Anatomy of an extensible open source PACS. J Digit Imaging, 2015. 10.1007/s10278-015-9834-0 [DOI] [PMC free article] [PubMed]
- 21.Costa C, Freitas F, Pereira M, Silva A, Oliveira JL. Indexing and retrieving DICOM data in disperse and unstructured archives. Int J Comput Assist Radiol Surg. 2009;4(1):71–77. doi: 10.1007/s11548-008-0269-7. [DOI] [PubMed] [Google Scholar]
- 22.Bastiao L, Santos M, Costa C, Silva A, Rocha N. Dicoogle statistics: Analyzing efficiency and service quality of digital imaging laboratories. Heildelberg: Springer; 2013. [Google Scholar]
- 23.Santos M, Bastiao L, Costa C, Silva A, Rocha N. Dicom and clinical data mining in a small hospital pacs: A pilot study. In: International Conference on ENTERprise Information Systems. Berlin: Springer; 2011. pp. 254–263. [Google Scholar]