Abstract
Regulated ion diffusion across biological membranes is vital for cell function. In a nanoscale ion channel, the active role of discrete water molecules in modulating hydrodynamic behaviors of individual ions is poorly understood because of the technical challenge of tracking water molecules through the channel. Here we report the results of a hydroxyl radical footprinting analysis of the zinc-selective channel ZIPB from the Gram-negative bacterium, Bordetella bronchiseptica. Irradiating ZIPB by microsecond X-ray pulses activated water molecules to form covalent hydroxyl radical adducts at nearby residues, which were identified by bottom-up proteomics to detect residues that interact either with zinc or water in response to zinc binding. We found a series of residues exhibiting reciprocal changes in water accessibility attributed to alternating zinc and water binding. Mapping these residues to the previously reported crystal structure of ZIPB, we identified a water-reactive pathway that superimposed on a zinc translocation pathway consisting of two binuclear metal centers and an interim zinc-binding site. The cotranslocation of zinc and water suggested that pore-lining residues undergo a mode switch between zinc coordination and water binding to confer zinc mobility. The unprecedented details of water-mediated zinc transport identified here highlight an essential role of solvated waters in driving zinc coordination dynamics and transmembrane crossing.
Keywords: zinc, transport, membrane protein, transport metal, protein conformation, metal ion–protein interaction, mechanism, water, X-ray footprinting
Introduction
Discrete water molecules in confined structures of membrane channels and transporters are expected to exhibit distinct physicochemical behaviors not observed in the bulk solvent conductive for free diffusion. Theoretical calculations suggested an essential role of solvated water molecules in shaping the selectivity and conductivity of a potassium channel, KcsA (1). However, a rapid potassium flux at a rate approaching the diffusion limit precluded experiments from tracking interactions between water molecules and channel-lining residues when the ion floodgate in KcsA is switched open. On the other hand, the rate of zinc flux in a zinc-selective transporter, ZIPB, is several orders of magnitude slower than expected for a typical ion channel (2). ZIPB is a prototypic zinc transporter in the ubiquitous Zrt-, Irt-like protein (ZIP)3 family (3). Earlier biochemical studies showed that zinc flux mediated by ZIPB was nonsaturable, electrogenic, and voltage-dependent (2). The zinc equilibrium potential exhibited a Nernst relationship, whereas the voltage dependence of the zinc flux also followed the Goldman–Hodgkin–Katz current equation (2). Thus, ZIPB-mediated zinc transport is a slow electrodiffusion process, providing a unique experimental system to investigate how solvated water molecules may interact with the channel wall to tune the binding and release of zinc ions when they pass through a zinc-specific channel.
The binding stabilities for zinc capture and release could differ by many orders of magnitude (4). The crystal structures of ZIPB in complex with Zn/Cd revealed an occluded translocation pathway defined by a sequence of five bound metal ions (5). A bound Zn and an isomorphous Cd ion in proximity are bridged by two invariant glutamate residues to form a binuclear metal center in the middle of the transmembrane domain. Additional coordinating ligands from surrounding residues and water molecules saturate the coordination sphere. The highly constrained Zn/Cd-coordinating system in ZIPB is similar to that trapping bound Zn ions in many metalloproteins, where Zn is generally considered a permanent constituent of catalytic or structural sites (6). It is not clear how the bound Zn and Cd are destabilized in ZIPB to render mobility. Of note, several crystallographic water molecules were found in the hydration shells of bound metal ions and also in close proximity to protein ligands that participated in binuclear metal coordination. Therefore, position shifts of certain coordinating residues via ZIPB conformational dynamics could switch interactions from a bound metal ion to solvation water molecules, favoring zinc release over binding. The possibility of alternating interactions with zinc and water molecules leads to the hypothesis that zinc and water molecules may be cotransported in ZIPB.
To identify a putative water translocation pathway in ZIPB, we used X-ray footprinting and mass spectrometry (XFMS) to detect regions of full-length ZIPB disposed toward either zinc or water interactions in response to zinc binding (7–9). The XFMS experiments were performed in aqueous solution, allowing protein conformational dynamics between a Zn-bound or Zn-free state. In the absence of zinc binding, ionizable residues in the low dielectric environment of the ZIPB interior are screened by water molecules to reduce electrostatic interactions. A common assumption is that Zn binding could expel water molecules from binding sites, reducing the water accessibility to coordinating residues. To track changes in water accessibility to individual residues in ZIPB, we used a high-flux-density X-ray pulse to generate hydroxyl radicals by in situ radiolysis of local or bound water molecules (9). The water-like van der Waals area and solvent properties of hydroxyl radicals are ideal as solvent accessibility probes (10, 11). The reactions between short-lived hydroxyl radicals and nearby residues yielded oxygen-based mass adducts to side chains (10). A ratiometric measurement of the rate constants in the presence and absence of zinc binding yielded an unbiased measure of water accessibility changes independent of the intrinsic residue reactivity (7, 9, 12).
Contrary to the expectation that Zn binding would block water access, we found reciprocal changes of water accessibilities to individual residues in subpockets of the binuclear metal center. Although a loss of water accessibility to certain residues was consistent with the protective effect of zinc binding, an increase in water accessibility following zinc binding indicated an influx of water molecules in association with the bound zinc ion. Mapping water-reactive residues to the ZIPB crystal structure revealed a water translocation pathway that overlapped a zinc translocation pathway defined by X-ray crystallography. Our analysis further revealed a route of water entry from one side of the binuclear metal center to disengage highly constrained metal ions, promoting their release and transmembrane movement.
Results
Radiolytic labeling and sequence coverage
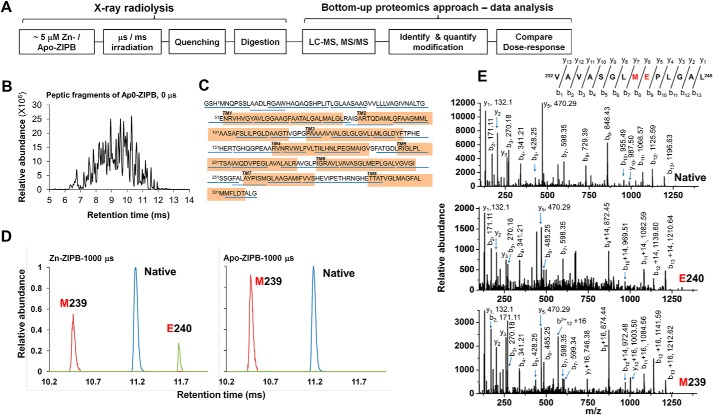
The high occupancy of the binuclear metal center in the crystal structure of ZIPB was enforced by an exceedingly high Zn or Cd ion concentration (100 mm) (5). A lower metal concentration was required to shift ZIPB to a more physiologically relevant conformation that allowed metal release. Here the zinc-induced conformational change was probed by quantifying changes in water accessibility to residues throughout the full-length ZIPB protein sequence. ZIPB in detergent micelles was stripped of residual metal ions by EDTA chelation, HPLC-purified, and then rapidly mixed with 0.2 mm ZnSO4, followed by a microsecond pulse of high-flux-density X-ray exposure to trigger hydroxyl radical labeling of reactive residues (Fig. 1A) (13). A progressive increase in X-ray exposure was applied, and the X-ray dosages were adjusted to a level so that the loss of unmodified peptides followed pseudo-first-order kinetics as a function of the exposure time (10). The irradiated samples were rapidly quenched by methionine amide to prevent secondary oxidation mediated by slow radical reactions (14). The resulting ZIPB samples were digested and then analyzed by high-flow LC-MS (Fig. 1B), which generated over 90% sequence coverage (Fig. 1C).
Figure 1.
Radiolytic labeling and high-resolution LC-MS analysis. A, experimental scheme. B, representative reverse-phase mass chromatogram. C, peptic fragments detected and confirmed by MS/MS analysis are underlined in blue. The transmembrane domains are highlighted in orange. D, extracted ion-count chromatograms of doubly protonated, native (11.2 min, 664.35 m/z, blue), carbonylated (11.7 min, +14-Da mass shift, 671.35 m/z, green), and hydroxylated (10.5 min, + 16-Da mass shift, 672.35 m/z, red) peptide 232–245 in Zn-ZIPB or apo-ZIPB, as indicated, each with a 1000-μs X-ray exposure. E, MS/MS identification of +14- and +16-Da modification of Glu240 and Met239, respectively.
Detection of radiolytic labeling at single-residue resolution
Sites of oxidative modifications that located on both transmembrane domains and periplasmic loops were identified from each modified peptide (Table 1), and similar extents of side-chain modifications were consistently observed at the same residues for repeated experiments. As shown in Fig. 1D, high-resolution LC-MS analysis of an exposed peptide, p232–245 (VAVASGLMEPLGAL) from zinc-bound ZIPB (Zn-ZIPB) resolved three species at distinct retention times, corresponding to a mass shift of +16 Da for the hydroxylated (10) and +14 Da for the carbonylated (10) and the unmodified products. In contrast, the same peptides from zinc-free ZIPB (apo-ZIPB) showed a hydroxylated and an unmodified peak. Comparing individual peak heights between two mass chromatograms revealed that zinc binding decreased hydroxylated modification, in contrast to a significant increase in carbonylated modification. The MS/MS assignments confirmed that modifications at Glu240 and Met239 yielded +14- and +16-Da mass shifts (Fig. 1E), identifying two consecutive residues with opposite radiolytic labeling responses to zinc binding.
Table 1.
The ratio of hydroxyl radical reactivity between Apo-ZIPB and Zn-ZIPB
| Sequence No.a | Peptide sequenceb | Site of modificationc | kApp-ZIPBd | kZn-ZIPBd | Re = kZn-ZIPB/kApo-ZIPB |
|---|---|---|---|---|---|
| (s−1) | (s−1) | ||||
| 2–6 | GSHMNQPSSL | Met1 (+16 Da)f | 335.7 ± 24 | 269.2 ± 23 | 0.81 ± 0.13 |
| 8–15 | AADLRGAW | Noneg | —g | — | — |
| 13–26 | RGAWHAQAQSHPLIT | His17 (+16 Da) | 13.2 ± 0.7 | 6.4 ± 1.5 | 0.52 ± 0.15 |
| 27–35 | LGLAASAAG | None | — | — | — |
| 39–58 | LLVAGIVNALTGENRVHVGY | None | — | — | — |
| 59–67 | AVLGGAAGF | None | — | — | — |
| 68–75 | AATALGAL | None | — | — | — |
| 76–82 | MALGLRA | None | — | — | — |
| 81–92 | RAISARTQDAML | None | — | — | — |
| 93–100 | GFAAGMML | Met98, Met99 (+16 Da) | 31.6 ± 4.3 | 28.6 ± 4.2 | 0.94 ± 0.26 |
| 106–113 | FSLILPGLD | None | — | — | — |
| 114–126 | AAGTIVGPGPAAA | None | — | — | — |
| 129–138 | VALGLGLGVL | None | — | — | — |
| 139–144 | LMLGLD | None | — | — | — |
| 145–153 | YFTPHEHER | Tyr145, Pro148, His149, Glu150 | 24.3 ± 2.7 | 2.7 ± 1.1 | 0.13 ± 0.01 |
| 154–164 | TGHQGPEAARV | His156 | 2.9 ± 0.05 | 0.05 | <0.05 |
| 165–170 | NRVWLF | None | — | — | — |
| 173–184 | TIILHNLPEGMA | Glu181, Met183 (+16 Da) | 37.9 ± 4.9 | 0.05 | <0.05 |
| 190–203 | ATGDLRIGLPLTSA | Asp193, Leu194 (+14 Da) | 2.8 ± 0.18 | 2.6 ± 0.18 | 0.94 ± 0.13 |
| 204–213 | IAIQDVPEGL | Modification unassigned | 0.05 | 7.1 ± 1.9 | > 20 |
| 217–231 | LALRAVGLPIGRAVL | +16 Da modification unassigned,g Leu219, Arg220, Ala221 (+14 Da) | 0.05 | 217.6 ± 9.6 | > 20 |
| 232–245 | VAVASGLMEPLGAL | Met239 (+16 Da) | 110 ± 11.3 | 60.8 ± 12.0 | 0.59 ± 0.12 |
| Glu240 (+14 Da) | 0.05 | 12.6 ± 1.1 | > 20 | ||
| 246–255 | VGVGISSGFA | None | — | — | — |
| 254–262 | FALAYPISM | None | — | — | — |
| 263–271 | GLAAGAMIF | Met269 (+16 Da) | 120 ± 12 | 0.05 | <0.05 |
| 272–291 | VVSHEVIPETHRNGHETTAT | His275, Glu276 (+16 Da) | 6.8 ± 2.8 | 30.0 ± 2.8 | 5.03 ± 2.4 |
| 302–309 | MFLDTALG | None | — | — | — |
a 94% sequence coverage was obtained from the bottom up LC–electrospray ionization–MS analysis of apo- and Zn-ZIPB using pepsin digestion.
b Sequences of digested fragments identified by mass spectrometry and used for the analysis as described under “Experimental procedures.”
c Positions of the most abundant modified residues identified by mass spectrometry analysis as described under “Experimental procedures.” Multiple residues are indicated by sequence number.
d The rate constant (k s−1) was estimated by employing a non-linear fit of hydroxyl radical modification data to a first-order decay as described under “Experimental procedures” (Fig 2).
e The ratio of the hydroxyl radical reactivity rate between Zn-ZIPB and apo-ZIPB. R is a quantitative measure of the change in solvent accessibility. The error of the R is the estimated lower and upper limit of the ratio based on the rate constant errors.
f Mass shift because of side-chain modification is shown in parentheses. Representative MS/MS plots are shown in Fig. 1E.
g No modification was detected, and no rate was determined.
Site-specific water accessibility change
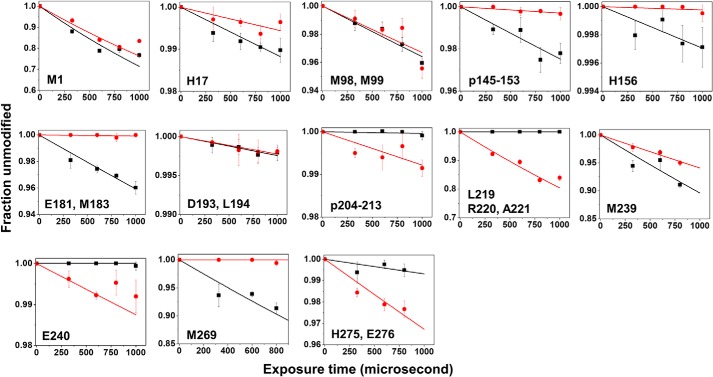
The extent of modification for each residue or peptide was calculated using the peak area of extracted ion counts and then plotted against a series of X-ray exposures to generate a site-specific dose–response plot (9). Pairwise comparison of dose plots for apo- and Zn-ZIPB revealed a zinc-dependent water accessibility change (Fig. 2). For example, comparing the dose–response plots for Met239 showed that zinc binding reduced hydroxyl radical labeling at higher X-ray dosages. In contrast, zinc binding greatly increased Glu240 labeling at higher dosages, demonstrating the reciprocal nature of the hydroxyl radical reactivities at two consecutive residues. The hydroxyl radical reactivity rate k (s−1) for each modified residue or peptide was calculated and summarized in Table 1.
Figure 2.
Hydroxyl radical reactivity. Dose–response plot for apo- ZIPB (black) or Zn-ZIPB (red) are shown for a specific residue or peptide as indicated. The solid lines indicate a single-parameter exponential fit as described under “Experimental procedures,” and the corresponding hydroxyl radical reactivity rate k (s−1) is summarized in Table 1.
Mapping water accessibility changes to the ZIPB crystal structure
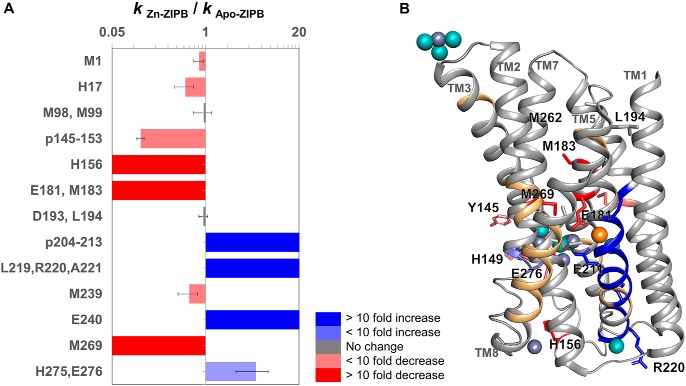
A ratiometric measurement of the rate constants (kZn-ZIPB/kapo-ZIPB) in the presence and absence of zinc binding yielded an unbiased measure of water accessibility change (Fig. 3A and Table 1) (7, 9). Zinc binding was found to induce protein conformational changes associated with a range of increases (r = 2 to >20) or decreases (r = 0.5 to <0.05) mapped to respective transmembrane helices (TMs) as well as extramembranous loops (Fig. 3B). Peptides not detected included less than 10% of the ZIPB sequences (Fig. 3B), and the residues in these regions did not contribute to zinc-binding sites in the crystal structure. The modification levels of residues within the hydrophobic region of ZIPB were less than 2% of the total residue or peptide signals, at a level similar to residue modifications in internal cavities of membrane proteins, where the water reactivity is governed by side-chain proximity to hydroxyl radicals originating from tightly bound water molecules (15).
Figure 3.
Mapping of water accessibility changes. A, the ratio (R) of hydroxyl reactivity rates as described in Table 1 for each residue or peptide shows decreased (red shades), increased (blue shades), or unchanged (gray) water accessibility in response to zinc binding. B, modified residues (sticks) and peptides (ribbons) in the crystal structure of Zn/Cd-ZIPB (PDB code 5TSA) (5) are colored according to the direction of water accessibility change as described in A. Peptides not detected are colored light orange. Bound Zn, Cd, and water molecules are shown as slate gray, orange, and cyan spheres, respectively.
Correlation between hydroxyl radical labeling and zinc binding
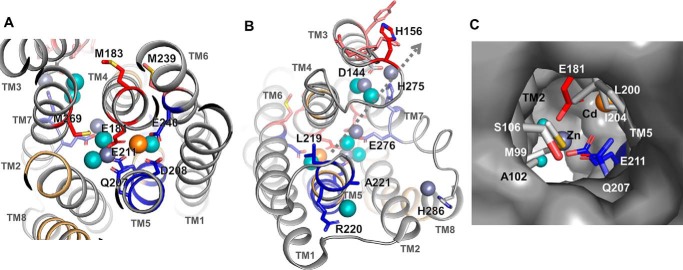
Among all the detectable proteolytic peptides generated by XFMS, zinc binding significantly protected four residues/peptides from hydroxyl radical labeling. The first group of completely protected residues was derived from peptide p173–184 (Table 1), including the invariant Glu181, which bridged two metal ions in a binuclear metal center located in the middle of the translocation pathway, and Met183 from TM4, which is packed against a portion of TM7 toward the extracellular side (Fig. 4A). The next highly protected cluster of residues was derived from peptide p145–153 and His156 from peptide p154–164 (Table 1). These two peptides formed a loop linking the cytoplasmic ends of TM3 and TM4 (Fig. 4B). The TM3-TM4 loop was displayed in an arbitrary conformation because it was not resolved in the crystal structure. Nevertheless, two confined zinc ions in the crystal structure were found in a cytoplasmic opening framed by the cytoplasmic ends of TM3, TM4, and TM7 (Fig. 4B). The bound zinc ions were bridged by an invariant Asp144 projected from TM3 and also coordinated with a conserved His275 from TM7 (Fig. 4B). The strong protection against hydroxyl radical modification to the TM3-TM4 loop by zinc binding indicated that the crystallographically unresolved loop was recruited to complete a second binuclear metal center on the cytoplasmic surface of ZIPB. The fourth protected residue, Met269, came from p263–271 (Table 1), which spanned the extracellular portion of TM7. Met269 in the crystal structure was packed against the portion of TM4 toward the extracellular side (Fig. 4A). The positioning of two fully protected residues (Met269 and Met183) at the TM7-TM4 interface indicated that zinc binding triggered a conformational change tightening interhelical packing on one side of the central binuclear metal center (Fig. 4A).
Figure 4.
Key residues involved in metal binding and water interactions. The residues are mapped in the crystal structure of ZIPB (5) and colored red and blue for decreased or increased water accessibility, as shown in Fig. 3. Bound Zn, Cd, and water molecules are shown as slate gray, orange, and cyan spheres, respectively. A, the central binuclear metal center viewed from the extracellular side. A pair of Zn and Cd ions is bridged by Glu181 and Glu211. Note that the clustering of modified residues with reciprocal changes in water accessibility is associated with crystallographic water molecules. B, the peripheral binuclear metal center viewed from the cytoplasmic side. The TM3-TM4 and TM7-TM8 loops are shown in arbitrary conformations. The dotted line indicates a plausible zinc translocation pathway from the central binuclear metal center through an intermediate zinc binding site at Glu276 toward the peripheral binuclear metal center near the cytoplasmic exit. C, extracellular entrance viewed approximately along the arrow, as indicated in B. Residues sealing the channel opening on the extracellular surface are drawn as sticks and excluded from the protein surface drawing.
Correlation between hydroxyl radical labeling and crystallographic water molecules
Zinc transport through ZIPB is accompanied by solvent influx into the translocation pathway. Four peptides were identified, with significant increases in hydroxyl radical labeling in response to zinc exposure. The first hyperreactive peptide, p204–213, harbored three coordination residues (Gln207, Asp208, and Glu211) in the middle of TM5 (Table 1). In the ZIPB crystal structure, these residues (Fig. 4A, blue) were on the same helical face, lining up one side of the binuclear metal center. Apparently, the crystallographic water molecules in the vicinity (within 3.5 Å, Fig. 4A) were shifted to enter the binuclear metal center along TM5. The second group of hyperreactive residues (Leu219, Arg220, and Ala221 from peptide p217–231; Table 1) identified an expanded water accessibility increase to a portion of TM5 toward the cytoplasmic side (Fig. 4B). In this region, a crystallographic water molecule was found in hydrogen bonding distance with the main-chain carbonyl of Leu219 and in proximity to the side chain of Arg220. A third hyperreactive peptide, p232–245 (Table 1), contained an invariant coordination residue, Glu240, in the middle of TM6 (Fig. 4A). This residue sealed a side of the binuclear metal center by coordinating the unexchangeable Cd ion in the crystal structure (Fig. 4A). A significant increase in water accessibility to Glu240 indicated that zinc binding altered the Glu240 coordination partner from Cd to a crystallographic water molecule. Consequently, the unexchangeable Cd site revealed in the ZIPB crystal structure became mobile in Cd-free ZIPB at a lower zinc concentration. Interestingly, water accessibility to the neighboring Met239 showed an opposite change with an ∼2-fold decrease (Fig. 4A). Met239 was packed against a peptide, p173–184, from TM4, where zinc binding was shown to protect it from labeling. The last hyperreactive peptide, p272–291 (Table 1), formed a cytoplasmic loop connecting the cytoplasmic ends of TM7 and TM8. This loop was unresolved in the crystal structure (Fig. 4B). It began with a conserved His275 at the cytoplasmic end of TM7, coordinating one of the two zinc ions in the peripheral binuclear metal center. An adjacent invariant Glu276 donated an oxygen ligand to another bound zinc located between the peripheral and central binuclear metal center. The increased rate of residue labeling was correlated with two crystallographic water molecules, each in close proximity to His275 or Glu276 (Fig. 4B). A third zinc ion associated with this hyperreactive loop was coordinated by a conserved His286 at the cytoplasmic end of TM8. Although no crystallographic water molecule was resolved in this region, this solvent-exposed area was susceptible to hydroxyl radical labeling from the bulk solvent. Of note, all three bound zinc ions were undercoordinated in the crystal structure. The missing protein ligands may be provided by coordination residues from unresolved TM3-TM4 and TM7-TM8 loops, which responded to zinc binding with opposite conformational changes in terms of decreased and increased water accessibility changes, respectively (Fig. 4B).
Water accessibility to the extracellular entrance
Among all detectable hydroxyl radical labels, only two reactive peptides showed no difference in water accessibility in response to zinc binding. Labeled residues in the first peptide included Asp193 and Leu194 (Fig. 3A), localized to the extracellular end of TM5 (Fig. 3B). Both residues were fully solvent-exposed. Two helical turns toward the inner membrane, Leu200 and Ile204 on the same helical face, were packed against Ala102 and Met99 from the opposing helical face of TM2 (Fig. 4C). In the crystal structure, these hydrophobic resides formed a tight seal that closed up the extracellular entrance to the metal translocation pathway (5). Met99/Met98 from the second reactive peptide, however, were accessible to hydroxyl radical labeling regardless of metal binding (Fig. 3A). The zinc-independent labeling of Met99/Met98 indicated no significant conformational change associated with the wetting behavior of the extracellular entrance. As observed previously, modifications of internal Met residues reflected local conformational flexibility (16). Water reactivities toward Met99/Met98 suggested that these buried residues in the ZIPB crystals became solvent-accessible. Thus, the closed extracellular entrance revealed in the crystal structure may be sufficiently dynamic for the passage of partially dehydrated metal ions at a slow rate, in agreement with a highly restricted electrodiffusional process through the ZIPB channel (2).
Discussion
Zinc transporters in the ZIP family constitute a major uptake route for dietary zinc acquisition. ZIPB was identified as a Zn/Cd-specific transporter and a structural biology target (2). A high binding affinity for zinc is a prerequisite for effective zinc capture by ZIPB (17), but the high-affinity may trap bound zinc to impede its transmembrane movement. The dynamic process of water access to the binuclear metal center revealed by XFMS uncovers an active role of hydration water molecules in releasing the trapped zinc ions. Remarkably, all water-reactive residues in the transmembrane region are invariant or highly conserved, suggesting that the functional roles of water interactions are well preserved in the ZIP protein family. The ordered water molecules revealed by the crystal structure are of sufficient occupancy to be detected by X-ray diffraction. They are frequently observed in transmembrane domains, and their interactions with polar or ionizable residues are among most conserved structural elements contributing to protein stability and conformational dynamics (18). Similar to a network of conserved water interactions lining a signal transmission channel in rhodopsins (15), water-reactive residues in ZIPB participate in zinc coordination in a sequence of zinc-binding sites (Fig. 5). Multiple zinc bindings concomitant with increased water accessibility to selective coordinating residues in each binding site suggest that zinc ions and water molecules are cotransported. In the central binuclear metal center, zinc coordination fully protected Glu181 from TM4, in agreement with a strong interaction with the bound zinc ion. In contrast, zinc binding increased water access to the Glu207-Asp208-Glu211 triad and Glu240 from TM5 and TM6, respectively (Fig. 5). The lack of zinc protection on the TM5/TM6 surface reflected uneven contributions of coordinating residues to zinc binding in the binuclear metal center (Fig. 4A). A relatively more dynamic zinc interaction on the TM5/TM6 helical surface may allow water entry to partially rehydrate a subpocket of the binuclear metal center, switching the binding mode of coordination residues to zinc release, as indicated by an increase in water reactivity.
Figure 5.
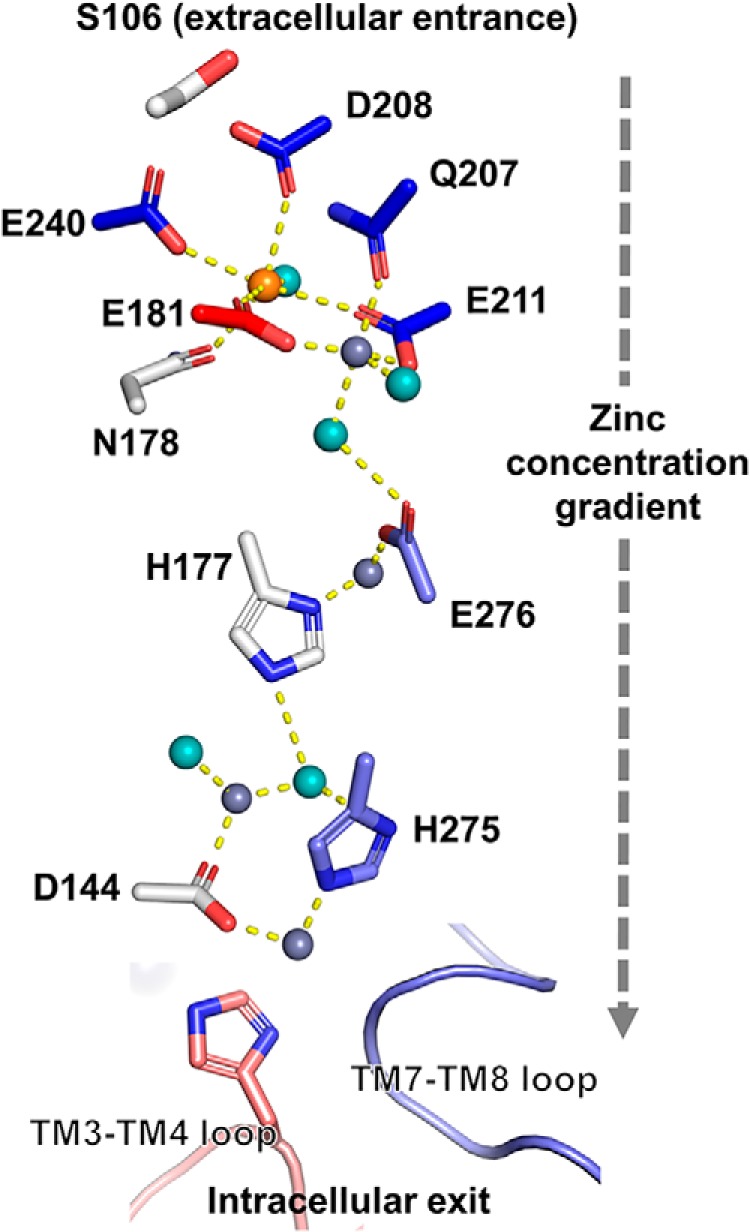
Schematic of water-mediated zinc transport. Multiple bound Zn/Cd ions in the crystal structure of ZIPB (5) define a zinc translocation pathway for zinc movement down a concentration gradient from an extracellular entrance marked by Ser106 to a cytoplasmic exit capped by two cytosolic loops not resolved in the crystal structure. Red, blue, and gray indicate protected, hyperreactive, and unmodified residues, as shown in Fig. 3. Bound Zn, Cd, and water molecules are shown as slate gray, orange, and cyan spheres, respectively. Yellow dotted lines show the distances, which are within 2.0–3.3 Å, between crystallographic water molecules, metal ions, and coordinating residues.
Following the central binuclear metal center, zinc ions navigate through the translocation pathway via an interim zinc-binding site to a peripheral binuclear metal center at the cytoplasmic exit (Fig. 5). These binding sites are closely spaced, allowing a series of ligand exchanges between consecutive binding sites to relay a bound zinc ion from one binding site to another down a zinc concentration gradient (19). Because there is virtually no free cytosolic zinc in cells because of a large excess of cellular zinc buffering capacity (20), the peripheral binuclear metal center is thought to form a high-affinity sink to hold imported zinc before the bound zinc is accepted by cytosolic zinc-binding proteins such as metallochaperones (21). Consequently, when zinc is available in the extracellular medium, an inward zinc concentration gradient would drive selective zinc diffusion in ZIPB (Fig. 5). This diffusional mechanism was similarly ascribed to the copper uptake transport Ctr1, where Cu+ diffusion is mediated by consecutive Cu+-binding sites, leading toward a high-affinity copper sink at the cytoplasmic exit (22). Finally, XFMS analysis revealed that two histidine-rich loops capping the cytoplasmic exit contribute additional coordination residues to complete the peripheral binuclear metal center (Fig. 5). Zinc binding protected the TM3-TM4 loop but exposed His275 from the cytoplasmic end of TM7, where the adjacent Glu276 also gained water accessibility upon zinc binding. His275 and Glu276 coordinate two different zinc ions and interact, respectively, with crystallographic water molecules within or near the hydrogen bonding distance, suggesting that the increased His275 and Glu276 reactivities are also derived from hydration water molecules of the moving zinc ions (Fig. 5).
In conclusion, the reciprocal pattern of water accessibility changes in both binuclear metal centers and their associations with vicinal crystallographic water molecules provide clear structural evidence for water-mediated movement of zinc ions along a sequence of five binding sites in ZIPB. In sharp contrast, a single tetrahedral zinc transport site in a proton-coupled zinc efflux transporter, YiiP (23), was completely protected from water access by zinc binding (7). Accordingly, zinc release in the transport cycle of YiiP is driven by conformational changes that alternatingly expose the transport site to either side of the membrane surfaces (7). Although mammalian ZIP transporters are thought to be operated by a similar alternating access mechanism (5, 24–26), our data from the bacterial ZIPB support a model in which water dynamics complement metal coordination chemistry to confer mobility to trapped zinc ions, highlighting the functional importance of solvated water in driving zinc transport through a sequence of high-affinity binding sites. Zinc is a defining feature of eukaryotic proteomes that are encoded by ∼10% of genomes (27). Biological zinc utilization depends on the trapping of zinc ions (28). Contrary to the expectation that zinc coordination would expel solvated water molecules, we found that zinc binding triggered water entry to ZIPB, revealing a different binding process to ensure zinc movement in and out of cells through zinc-specific channels and transporters.
Experimental procedures
Preparation of apo-ZIPB and Zn-ZIPB
His-tagged ZIPB was overexpressed in BL21(DE3) pLysS cells and purified as described previously (3). Briefly, cells in the stationary phase of an overnight autoinduction culture were harvested and lysed mechanically by three passages through a microfluidizer press cell. The resulting membrane vesicles were pelleted by ultracentrifugation and then solubilized using a detergent buffer containing 100 mm NaCl, 20 mm HEPES (pH 7.5), 7% dodecyl maltoside, 0.25 mm tris(2-carboxyethyl)phosphine (TCEP), and 20% (w/v) glycerol. The crude membrane extract was applied to a nickel-nitrilotriacetic acid Superflow column. After washing the column with 20 mm HEPES (pH 7.5), 300 mm NaCl, 20% (w/v) glycerol, 0.05% DDM, 0.25 mm TCEP, and 30 mm imidazole, ZIPB was eluted with an elevated imidazole concentration at 300 mm. The purified His-ZIPB was loaded to a 10-kDa-cutoff dialysis cassette against a bulk solution containing 20 mm HEPES (pH 7.5), 100 mm NaCl, 20% (w/v) glycerol, 0.05% DDM, and 0.25 mm TCEP. Thrombin (Novagen) was added to the dialysis cassette at a ratio of 1 unit/1 mg of His-ZIPB. The proteolytic removal of the His tag was confirmed by immunoblotting with an anti-His tag antibody. Prior to X-ray exposure, ZIPB was demetallized by incubation with 5 mm EDTA for 30 min. The EDTA-treated sample was then subjected to size-exclusion purification using a TSK 3000SWXL column pre-equilibrated with a radiolytic labeling buffer (10 mm NaPi (pH 6.5), 100 mm NaCl, and 0.02% DDM). The purified ZIPB is referred to as apo-ZIPB hereafter. The absence of bound zinc in apo-ZIPB was confirmed by synchrotron radiation X-ray fluorescence spectrometry. ZnCl2 was added to 0.2 mm to an aliquot of apo-ZIPB to form Zn-ZIPB.
Synchrotron X-ray radiolysis
The apo- and Zn-ZIPB samples at 5 μm concentration were exposed to high flux density X-ray using a standard microfluidic setup at the Advanced Light Source beamline 5.3.1. X-ray beam parameters were optimized to provide a sufficient dose of hydroxyl radicals in a submillisecond timeframe, as judged by a standard fluorophore assay. For each exposure, a fixed volume of a 200-μl sample in a 200-μm I.D. (internal diameter) Polymicro® capillary tube was passed through an X-ray window (200 × 500 μm2), and the flow rate was adjusted to yield an X-ray exposure time ranging from 250–1200 μs. All irradiations were carried out at 4 °C. The unwanted secondary oxidations in exposed samples were immediately quenched with 10 mm methionine amide and stored at −80 °C for LC-MS analysis. The “zero” sample was run under the same condition without opening the beamline shutter.
LC-MS
The exposed samples were subjected to Cys alkylation and detergent removal by cold acetone precipitation before 60-min pepsin digestion at pH 2 and 37 °C. Proteolyzed samples were analyzed in an Agilent 6550 iFunnel Q-TOF mass spectrometer (Agilent Technologies, Santa Clara, CA) coupled to an Agilent 1290 LC system (Agilent) using a Sigma-Aldrich Ascentis Peptides ES-C18 reverse phase column (2.1 × 100 mm, 2.7-μm particle size; Sigma-Aldrich, St. Louis, MO). Approximately 10 pmol of samples were loaded onto the column via an Infinity Autosampler (Agilent) with buffer A (2% acetonitrile and 0.1% formic acid) flowing at 0.400 ml/min. The peptides were separated and eluted into the mass spectrometer via a gradient with an initial condition of 5% buffer B (98% acetonitrile and 0.1% formic acid) increasing to 70% buffer B over 14 min. Subsequently, buffer B was increased to 90% over 1 min and held for 3 min at a flow rate of 0.6 ml/min, followed by a ramp back down to 5% over 1 min, where it was held for minutes to re-equilibrate the column to the original condition. Peptides were introduced to the mass spectrometer from the LC using a Jet Stream electrospray ionization source (Agilent) operating in positive ion mode (3500 V). We used a newly established standard flow LC method with a high degree of chromatographic reproducibility and increased throughput for sample analysis (12). For all exposures (0–1200 ms), we collected full electrospray ionization–MS or MS1 (300–1400 m/z)) data with MassHunter B.05.00. For the 1000- and 1200-ms exposures, the data were acquired under Auto MS/MS (MS2) mode, whereby the three most intense ions (charge states 2–5) within the 300–1400 m/z mass range above a threshold of 1000 counts were selected for MS/MS data acquisition. Mass spectra were collected with the quadrupole set to “narrow” resolution and collision energy to optimize fragmentation. MS/MS spectra were scanned from m/z 100–1700 and were collected until reaching 40,000 total counts or for a maximum accumulation time of 333 ms. Parent ions were excluded for 0.1 min following MS/MS acquisition. MS/MS data of native and modified peptide fragments were interpreted by Mascot MS/MS Ions Search as well as manual validation. Both MS1 and MS2 were run under the same LC gradient condition.
Identification of native and modified residues by MS/MS assignments
The MS/MS data were used for peptide sequence determination and identification of the position of modification by MASCOT automated MS/MS assignment. All MS/MS assignments were verified manually with the aid of Protein Prospector (version 5.21.2, University of California, San Francisco). The MS1 data were used for retention time–based peak area analysis to calculate the extent of native and modified peptides at any exposure time point using Agilent Mass Hunter v 2.0. All peaks and MS/MS assignments were manually verified using four basic characteristics of unmodified and modified products: accurate m/z, isotopic m/z distribution of the m/z of various charge states, retention time, and MS/MS sequencing. In cases where the MS/MS assessment was not available, the first three criteria were used for peptide and peak assignment, and only the overall modification of the peptide was reported.
Quantification of hydroxyl radial labeling
For apo- and Zn-ZIPB, the peak areas from the extracted ion chromatograms of a specific peptide fragment with a particular m/z and associated +16- and +14-Da side-chain modifications were used to quantify the amount of modifications at a given irradiation time. Increasing irradiation (0, 250, 500, 750, and 1000 μs) progressively reduced the fraction of unmodified products and provided a residue- or peptide-specific dose–response plot as shown in Fig. 2. All data were presented as mean ± S.D. based on two independent sample repeats and technical replicates. The hydroxyl radical reactivity rate (k s−1), which depends on both intrinsic reactivity and solvent accessibility, was obtained by fitting the dose response to a single exponential decay function: y = e−kx (based on a pseudo-first-order reaction scheme using Origin 7.5 (OriginLabs, Northampton, MA). The fitting results determined the reported errors of the labeling rate, as shown in Table 1. The deviation from linearity observed at high exposure was due to secondary radiation effects; hence, in some cases, the highest exposure data point (1000 μs) was not included in the curve fitting. The ratio (R) of the measured reactivity between Zn- and apo-ZIPB (R = kZn-ZIPB/kapo-ZIPB) yielded a quantitative measure of solvent accessibility changes independent of the intrinsic reactivity (Table 1).
Author contributions
S. G., C. M., and D. F. conceptualization; S. G., C. M., C. J. P., C. Y. R., and D. F. formal analysis; S. G., C. M., C. J. P., C. Y. R., and D. F. investigation; S. G., C. M., C. J. P., C. Y. R., and D. F. methodology; S. G., C. M., C. Y. R., and D. F. writing-original draft; S. G., C. M., C. Y. R., and D. F. writing-review and editing; C. J. P. and C. Y. R. resources; D. F. supervision; D. F. funding acquisition.
Acknowledgments
We thank Lean-Jade G. Chan for MS data collection at the Joint BioEnergy Institute. We thank Jun Feng for beamline support at beamline 5.3.1 of the Advanced Light Source. XFMS was conducted at the Advanced Light Source and Joint BioEnergy Institute, supported by the Office of Science, Office of Biological and Environmental Research of the Department of Energy under contract DE-AC02-05CH11231.
This work is supported by National Institutes of Health Grants 1R01GM126218 and 5R01DK108599. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ZIP
- Zrt-, Irt-like protein
- XFMS
- X-ray footprinting and MS
- TM
- transmembrane
- TCEP
- tris(2-carboxyethyl)phosphine
- DDM
- dodecyl maltoside.
References
- 1. Ostmeyer J., Chakrapani S., Pan A. C., Perozo E., and Roux B. (2013) Recovery from slow inactivation in K+ channels is controlled by water molecules. Nature 501, 121–124 10.1038/nature12395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lin W., Chai J., Love J., and Fu D. (2010) Selective electrodiffusion of zinc ions in a Zrt-, Irt-like protein, ZIPB. J. Biol. Chem. 285, 39013–39020 10.1074/jbc.M110.180620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jeong J., and Eide D. J. (2013) The SLC39 family of zinc transporters. Mol. Aspects Med. 34, 612–619 10.1016/j.mam.2012.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maret W. (2012) New perspectives of zinc coordination environments in proteins. J. Inorg. Biochem. 111, 110–116 10.1016/j.jinorgbio.2011.11.018 [DOI] [PubMed] [Google Scholar]
- 5. Zhang T., Liu J., Fellner M., Zhang C., Sui D., and Hu J. (2017) Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway. Sci. Adv. 3, e1700344 10.1126/sciadv.1700344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maret W., and Li Y. (2009) Coordination dynamics of zinc in proteins. Chem. Rev. 109, 4682–4707 10.1021/cr800556u [DOI] [PubMed] [Google Scholar]
- 7. Gupta S., Chai J., Cheng J., D'Mello R., Chance M. R., and Fu D. (2014) Visualizing the kinetic power stroke that drives proton-coupled zinc(II) transport. Nature 512, 101–104 10.1038/nature13382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gupta S., Bavro V. N., D'Mello R., Tucker S. J., Vénien-Bryan C., and Chance M. R. (2010) Conformational changes during the gating of a potassium channel revealed by structural mass spectrometry. Structure 18, 839–846 10.1016/j.str.2010.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gupta S., Feng J., Chan L. J., Petzold C. J., and Ralston C. Y. (2016) Synchrotron X-ray footprinting as a method to visualize water in proteins. J. Synchrotron Radiat. 23, 1056–1069 10.1107/S1600577516009024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu G., and Chance M. R. (2007) Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 107, 3514–3543 10.1021/cr0682047 [DOI] [PubMed] [Google Scholar]
- 11. Gupta S., D'Mello R., and Chance M. R. (2012) Structure and dynamics of protein waters revealed by radiolysis and mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 109, 14882–14887 10.1073/pnas.1209060109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. González Fernández-Niño S. M., Smith-Moritz A. M., Chan L. J., Adams P. D., Heazlewood J. L., and Petzold C. J. (2015) Standard flow liquid chromatography for shotgun proteomics in bioenergy research. Front. Bioeng. Biotechnol. 3, 44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gupta S., Celestre R., Petzold C. J., Chance M. R., and Ralston C. (2014) Development of a microsecond X-ray protein footprinting facility at the Advanced Light Source. J. Synchrotron. Radiat. 21, 690–699 10.1107/S1600577514007000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu G., Kiselar J., He Q., and Chance M. R. (2005) Secondary reactions and strategies to improve quantitative protein footprinting. Anal. Chem. 77, 3029–3037 10.1021/ac048282z [DOI] [PubMed] [Google Scholar]
- 15. Angel T. E., Gupta S., Jastrzebska B., Palczewski K., and Chance M. R. (2009) Structural waters define a functional channel mediating activation of the GPCR, rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 106, 14367–14372 10.1073/pnas.0901074106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zheng X., Wintrode P. L., and Chance M. R. (2008) Complementary structural mass spectrometry techniques reveal local dynamics in functionally important regions of a metastable serpin. Structure 16, 38–51 10.1016/j.str.2007.10.019 [DOI] [PubMed] [Google Scholar]
- 17. Roux B., Bernèche S., Egwolf B., Lev B., Noskov S. Y., Rowley C. N., and Yu H. (2011) Ion selectivity in channels and transporters. J. Gen. Physiol. 137, 415–426 10.1085/jgp.201010577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Renthal R. (2008) Buried water molecules in helical transmembrane proteins. Protein Sci. 17, 293–298 10.1110/ps.073237508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Banci L., Bertini I., Ciofi-Baffoni S., Kozyreva T., Zovo K., and Palumaa P. (2010) Affinity gradients drive copper to cellular destinations. Nature 465, 645–648 10.1038/nature09018 [DOI] [PubMed] [Google Scholar]
- 20. Outten C. E., and O'Halloran T. V. (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292, 2488–2492 10.1126/science.1060331 [DOI] [PubMed] [Google Scholar]
- 21. O'Halloran T. V., and Culotta V. C. (2000) Metallochaperones, an intracellular shuttle service for metal ions. J. Biol. Chem. 275, 25057–25060 10.1074/jbc.R000006200 [DOI] [PubMed] [Google Scholar]
- 22. De Feo C. J., Aller S. G., Siluvai G. S., Blackburn N. J., and Unger V. M. (2009) Three-dimensional structure of the human copper transporter hCTR1. Proc. Natl. Acad. Sci. U.S.A. 106, 4237–4242 10.1073/pnas.0810286106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu M., and Fu D. (2007) Structure of the zinc transporter YiiP. Science 317, 1746–1748 10.1126/science.1143748 [DOI] [PubMed] [Google Scholar]
- 24. Antala S., Ovchinnikov S., Kamisetty H., Baker D., and Dempski R. E. (2015) Computation and functional studies provide a model for the structure of the zinc transporter hZIP4. J. Biol. Chem. 290, 17796–17805 10.1074/jbc.M114.617613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Franz M. C., Pujol-Giménez J., Montalbetti N., Fernandez-Tenorio M., DeGrado T. R., Niggli E., Romero M. F., and Hediger M. A. (2018) Reassessment of the Transport mechanism of the human zinc transporter SLC39A2. Biochemistry 57, 3976–3986 10.1021/acs.biochem.8b00511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gyimesi G., Albano G., Fuster D. G., Hediger M. A., and Pujol-Giménez J. (2019) Unraveling the structural elements of pH sensitivity and substrate binding in the human zinc transporter SLC39A2 (ZIP2). J. Biol. Chem. 294, 8046–8063 10.1074/jbc.RA118.006113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Andreini C., Banci L., Bertini I., and Rosato A. (2006) Counting the zinc-proteins encoded in the human genome. J. Proteome Res. 5, 196–201 10.1021/pr050361j [DOI] [PubMed] [Google Scholar]
- 28. Vallee B. L., and Auld D. S. (1990) Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry. 29, 5647–5659 10.1021/bi00476a001 [DOI] [PubMed] [Google Scholar]