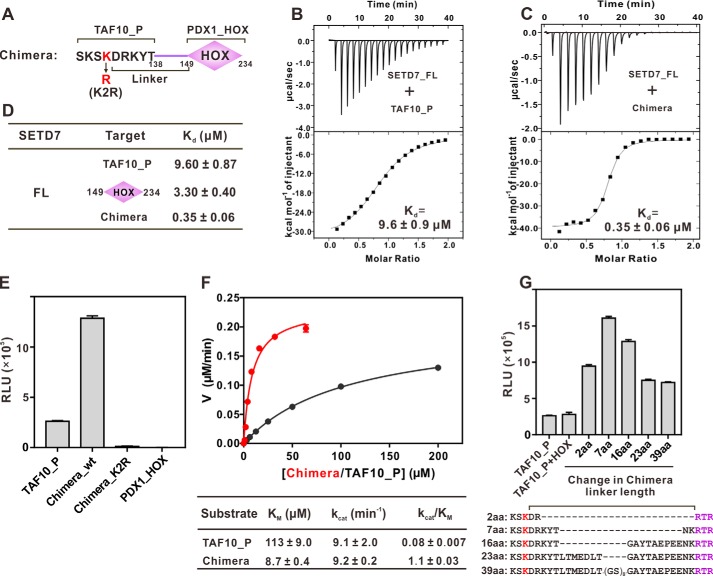
Figure 3.
Docking-induced enhancement of TAF10 methylation by SETD7. A, schematic diagram illustrating a chimera composed of TAF10 peptide (TAF10_P) fused with PDX1_HOX. The linker is defined as residues between the methylating Lys and Arg-149 of PDX1. B and C, ITC results quantifying the binding affinities of SETD7 to TAF10_P (B) and the chimera (C). D, ITC results summarizing the binding affinities of SETD7 to TAF10_P, PDX1_HOX, and chimera. E, in vitro assay of TAF10_P or chimera methylation by SETD7. The concentration of SETD7 used in the assay was 0.025 μm. F, Michaelis–Menten plots comparing the methylation kinetics of SETD7-mediated methylations of TAF10_P and the chimera. G, linker length between methylation site and docking site affects substrate methylation efficiency by SETD7. The detailed linker sequences are indicated in the bottom panel. For E and G, error bars represent the S.D. of three different batches of experiments.