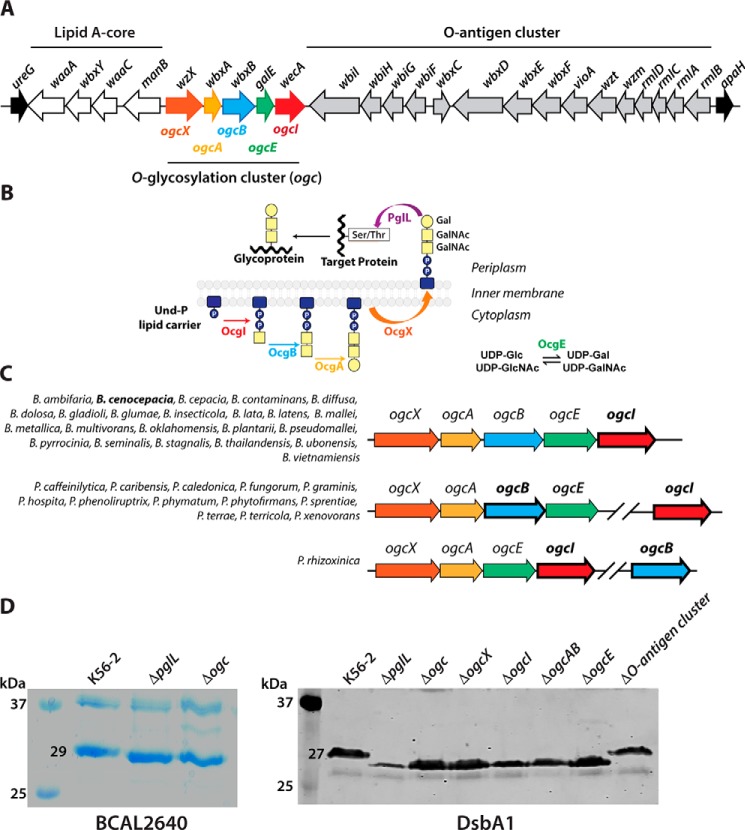
Figure 1.
Identification of the O-glycosylation cluster (ogc) in B. cenocepacia. A, genetic organization of the ogc cluster (BCAL31140–BCAL3118), placed between the O-antigen and lipid A-core clusters. Names of ORFs indicated above the arrows are according to the original gene annotation (25). Names below are the ones assigned in this study. B, proposed model for the general O-glycosylation assembly pathway in B. cenocepacia; OgcI, initiating enzyme; OgcE, UDP-glucose/galactose epimerase; OgcAB, glycosyltransferases; OgcX, flippase; and PglL, oligosaccharyltransferase. C, synteny of ogc genes in members of the Burkholderia genus. Genes indicated in bold are the ones used to search for synteny in the SyntTax server. The species showing similar arrangements are listed for each group. The synteny scores ranged from 55 to 96 (>30 is considered highly significant conservation) (26). D, relative gel mobility of BCAL2640 and DsbA1 polypeptides in ΔogcX–ogcI and single deletion mutants. Similar results were obtained in three biological repeats. Left panel, Coomassie-stained 14% SDS-polyacrylamide gel of the purified B. cenocepacia BCAL2640 protein expressed in WT K56-2, ΔpglL, and Δogc. Right panel, Western blotting of His-tagged DsbA1 acceptor protein expressed in the parental K56-2 strains and the mutants ΔpglL, ΔogcX, ΔogcX, ΔogcI, ΔogcAB, ΔogcE, and ΔO-antigen cluster (ΔBCAL3119–BCAL3131).