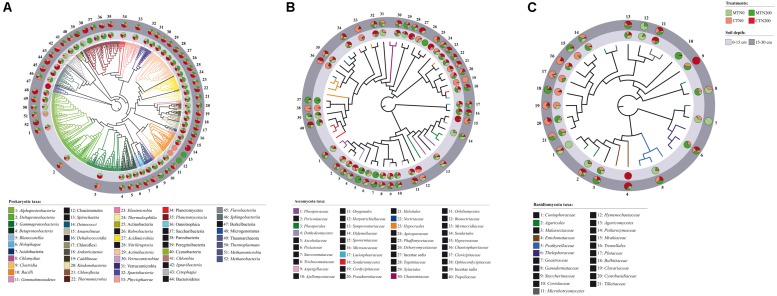
FIGURE 1.
Neighbor-joining (NJ) tree of 52 prokaryotic taxon representative sequences (A) and Maximum Likelihood (ML) trees of 40 Ascomycota (B) and 21 Basidiomycota taxon representative sequences (C) found in soil under long-term tillage and nitrogen fertilization. Treatments are: MTN0 (minimum tillage and 0 kg N ha–1), MTN200 (minimum tillage and 200 kg N ha–1), CTN0 (conventional tillage and 0 kg N ha–1) and CTN200 (conventional tillage and 200 kg N ha–1). Soil depths are: 0–15 and 15–30 cm soil depths. NJ tree of prokaryotes is based on the sequences obtained from the amplification of the V4 region (16 SSU rRNA gene) and ML trees of fungi are based on the sequences obtained from the amplification of the ITS1 region (for details see Supplementary Table S1). The prokaryotic taxa were assigned to Operational Taxonomic Unit (OTU) by BLAST against the 16S SSU SILVA database by clustering sequence reads at the 97% similarity threshold. The fungal taxa were assigned by BLAST against the ITS UNITE database and were selected by dynamic threshold values of clustering. For each OTU, the proportion of sequences retrieved from treatment (MTN0, light green; MTN200, dark green; CTN0, light red; CTN200, dark red) and soil depths (0–15 cm: light gray; 15–30 cm: dark gray) are shown in the pie charts. The name of each OTU is composed by the name of the class/phylum phylogenetic resolution and a serial number that is reported also in the trees.