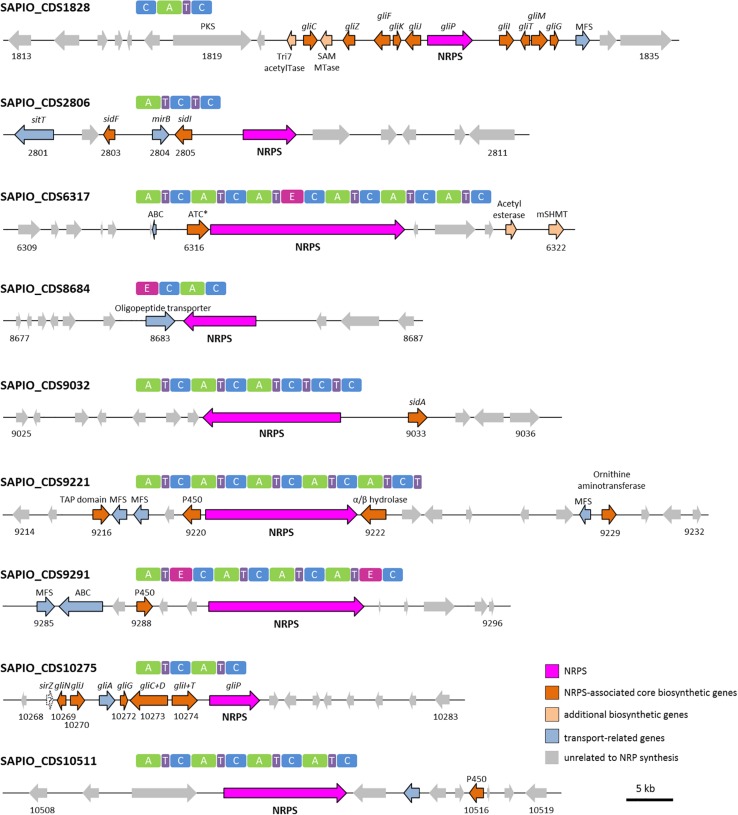
FIGURE 1.
NRPS genes and their domain architecture in Scedosporium apiospermum. Gene clusters shown are putatively involved in the biosynthesis of epidithiodioxopiperazines (CDS1828 and 10275), siderophores (CDS2806 and 9032), cyclopeptides (CDS6317 and 10511), and other unassigned derivatives (CDS8684, 9221 and 9291). All S. apiospermum NRPS genes are represented by pink/purple arrows. The minimal core module of an NRPS consists in an adenylation (A) domain for selection and activation of amino acids, a condensation (C) domain for catalyzing the formation of peptide bonds and a thiolation (T) domain for binding the amino acid building blocks. In addition to these standard modules, further structural variation can be introduced by optional domains such as epimerization (E) domains, which are responsible for conversion of L- to D-amino acids. Abbreviations: ABC, ATP binding-cassette; ATC∗, A-T-C module with truncated condensation domain; MFS, Major Facilitator Superfamily; P450, cytochrome P450 monooxygenase; mSHMT, mitochondrial serine hydroxymethyltransferase; SAM MTase, S-adenosyl-L-methionine-dependent methyltransferase; TAP, TAP-domain containing protein; Tri7 acetylTase, trichothecene 4-O-acetyltransferase.