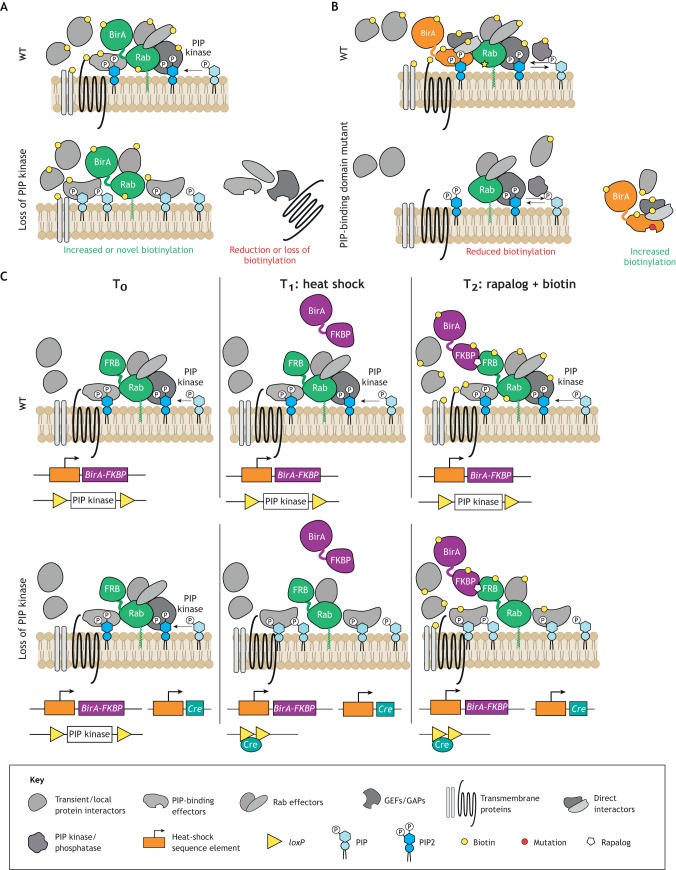
Fig. 3.
Potential experimental approaches to interrogate pleiotropy in phosphoinositide biology. PIPs and their effectors have pleiotropic functions. Elucidating these will require methods that capture global changes that occur when PIPs are altered. This figure shows the complex protein networks that interact with PIPs and illustrates the possible combination of BioID screening (Roux et al., 2013) with inducible protein expression/localization methods to investigate how PIP kinase mutants or PIP-binding protein mutants acquire differential subcellular protein interactomes. (A) A Rab GTPase can be used as an organelle marker that can be fused with the biotin protein ligase BirA, which biotinylates nearby endogenous PIP-interacting proteins, to provide subcellular specificity to a differential interactome. This interactome, mapped based on biotinylation (yellow circle), is different between wild-type cells (upper panel) and cells that do not express, for example, a PIP kinase (lower panel). (B) If BirA is fused to a PIP-binding protein, mutations in a PIP-binding protein (red circle) alter the interactome. This results in a BioID screening output showing biotinylation of PIP-independent interactors, whereas the PIP-containing compartment will lose or have reduced biotinylation. Although useful as an organelle marker, fusing Rab to BirA may result in BioID screens identifying non-specific transient interactions, such as those with the Rab GDP dissociation inhibitor GDI. These methods could be extended upon with artificial regulation of protein complex formation, such as the rapamycin- or rapalog-induced dimerization between FKBP- and FRB-fused proteins (Inobe and Nukina, 2016), and with inducible gene editing to increase spatiotemporal control. (C) Concomitant heat-shock-induced expression of the Rab-FRB fusion, the BirA-FKBP fusion and Cre recombinase, and addition of a rapalog to induce FKBP-FRB dimerization, allows for simultaneous control over protein biotinylation and Cre-induced deletion of a PIP kinase. This would allow for identification of differential subcellular protein interactomes before and shortly after knockout of a gene (in this example, a PIP kinase).