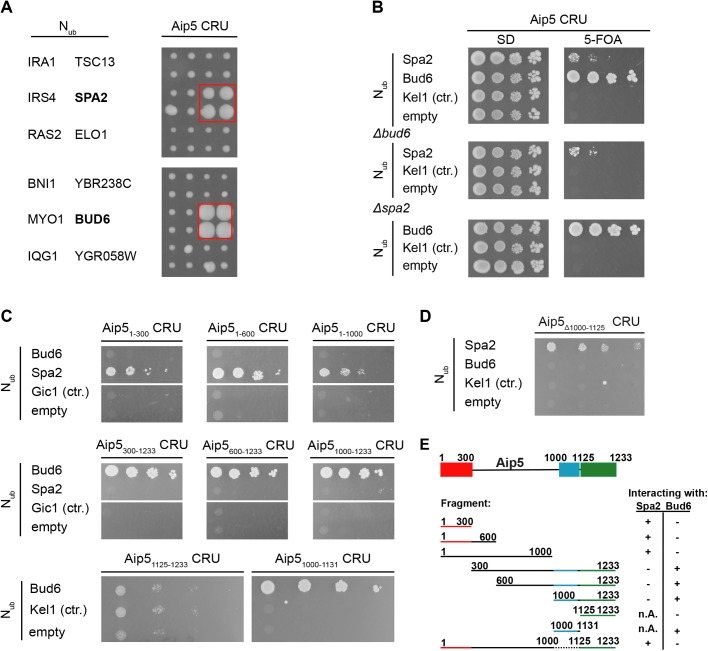
Fig. 1.
Aip5 interacts with Bud6 and Spa2 in vivo. (A) Right panel shows the cut outs of a split-Ubiquitin analysis of 533 diploid Aip5 CRU-containing yeast strains, each co-expressing a different Nub fusion protein. The Nub and CRU-expressing cells were independently mated four times, spotted in quadruplets, transferred onto medium containing 5-FOA and incubated for 4 days. Growth of four colonies indicates interaction. Left panel identifies the Nub fusions of the corresponding cut outs. Nub-Bud6- and Nub-Spa2 expressing cells are boxed (see also Fig. S1 for the complete array). (B) Bud6 and Spa2 interact independently of each other with Aip5. Haploid yeast strains (upper panel) lacking either BUD6 (middle panel) or SPA2 (lower panel) and expressing the indicated Nub fusions were spotted in tenfold serial dilutions starting at OD600=1 on medium containing (right panel) or not containing (left panel) 5-FOA. Cells were incubated for 7 days. (C) Split-Ub analysis as in B but with diploid yeast cells expressing full length Aip5 or fragments of Aip5 as CRU fusions together with the indicated Nub fusions. (D) Split-Ub assay as in B but with diploid yeast cells expressing genomically integrated Aip5Δ1000-1125 CRU and co-expressing the indicated Nub fusions. (E) Summary of the binding-site analysis. Spa2 binding site (red), Bud6 binding site (blue) and the predicted Grx-like domain (green) are highlighted in colors.