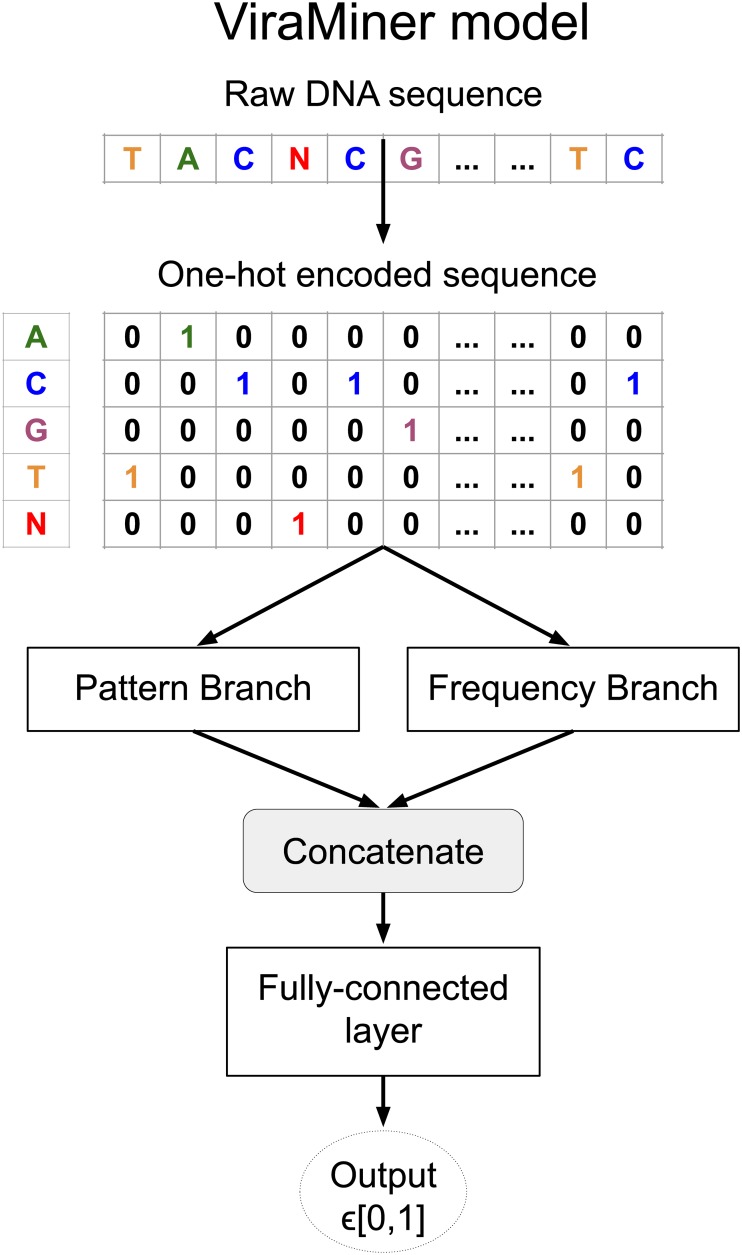
Fig 1. ViraMiner architecture.
ViraMiner architecture takes as input the raw sequences in one-hot encoded form. The raw sequences are then processed by two different convolutional branches (pattern and frequency branch). The architecture of the branches is shown in detail in Methods and Material. The outputs of the branches (two 1D vectors) are concatenated. This concatenated vector is all-to-one connected (fully-connected) with the output node. The output node passes the weighted sum of its inputs through sigmoid activation function resulting in a value restricted to range [0, 1]. This value reflects the likelihood of the sequence belonging to virus class.