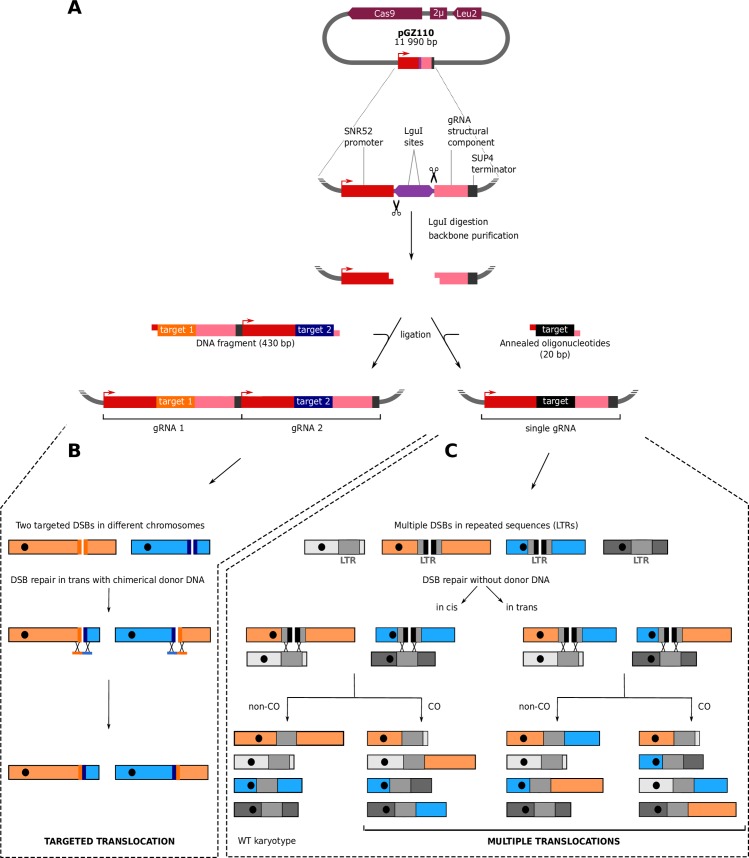
Fig 1. Strategy to reshuffle the yeast genome.
A. Cloning gRNA target sequences in the pGZ110 vector (Bruce Futcher, Stony Brook University). Upon digestion, the two LguI (also called SapI) sites generate non-complementary single strand overhangs of 3 bases. This system is versatile as it allows to clone in a single ligation step either a DNA fragment of 430 base-pairs reconstituting two different gRNA expression cassettes in tandem (left) or a 20 bp oligonucleotide corresponding to the target sequence of a unique gRNA (right). B. Induction of a targeted reciprocal translocation. Two DSBs are induced in different chromosomes and are repaired by homologous recombination with chimerical donor DNA oligonucleotides. The bold lines flanking the DSBs symbolize the target sequences. Donor DNAs force the repair to occur in trans (i.e. between two ends coming from the two different DSBs). C. Induction of multiple reciprocal translocations. This example illustrates a situation where the only two LTRs located in the blue and orange chromosomes are targeted by the gRNA (bold black lines indicate the target sequences) while LTRs located in the two grey chromosomes are not cut (devoid of target sequence). Note that more LTRs can be targeted as detailed below (see Fig 5). DSBs are repaired by homologous recombination using the uncut LTRs as donor template. Repair can occur in cis (i.e. the two ends from the same DSB are repaired together) or in trans (i.e. two ends from two different DSBs are repaired together).