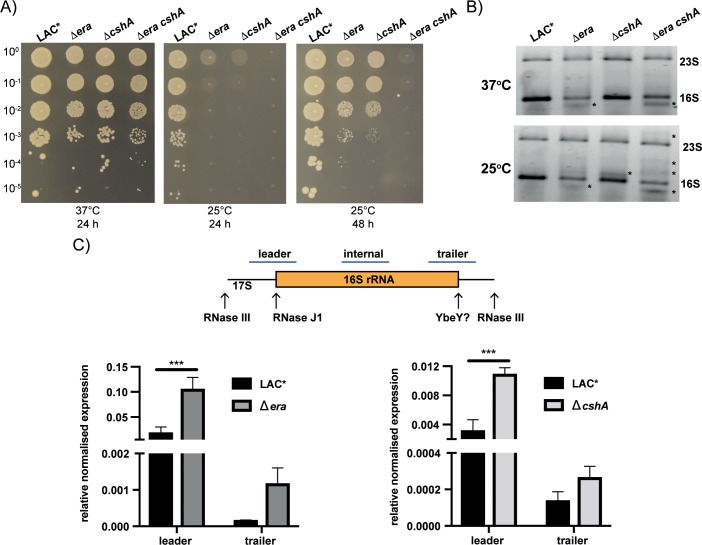
Fig 4. Era and CshA are required for cold adaptation and rRNA homeostasis.
A) Serial dilutions of the wildtype LAC*, Δera, ΔcshA and Δera cshA were spotted onto TSA agar plates and incubated at either 37 or 25°C for the times indicated. N = 3 with one representative image shown. B) rRNA profiles from wildtype, era and cshA mutant strains. 500 ng of RNA extracted from LAC*, Δera, ΔcshA and Δera cshA grown to an OD600 of 0.4 at either 37 or 25°C, were run on 0.7% agarose/0.9% synergel gels and stained with Evagreen dye. * highlights the presence of either processing or degradation intermediates. N = 3 with one representative image shown. C) top: schematic of the 17S rRNA, with arrows indicating processing sites that lead to the formation of the mature 16S rRNA. The blue bars indicate the probe regions amplified by RT-PCR. Bottom: RT-PCR of total RNA from the wildtype LAC*, Δera, and ΔcshA strains grown at 25°C using probes as indicated above. rRNA values between strains are normalised to the expression of rho RNA as an internal control before being plotted as mean expression +/- SD relative to the total 16S (internal probe set to a value of 1). The rho transcript was chosen as it has been shown to be highly stable under nutrient-deficient conditions under which the stringent response would be induced [38]. Statistical analysis was performed using a two-way ANOVA, followed by Sidak’s multiple comparisons test (*** P < 0.005). Cycle threshold values were determined for 3 biological repeats in duplicate (N = 3).