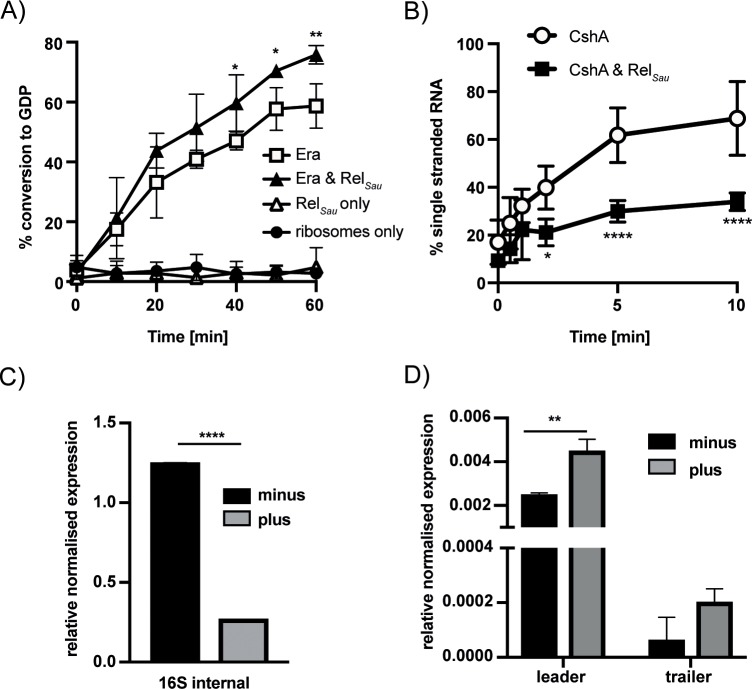
Fig 6. The stringent response affects Era and CshA enzyme activity and rRNA processing.
A) The GTPase activity of 100 nM Era was measured in the presence of an equal amount of ribosomes, 100 nM RelSau and 1 μM GTP. All reactions contained ribosomes. The RelSau only control reaction contained RelSau and ribosomes but no Era. Hydrolysis of 32P-GTP was monitored by TLC and the percentage GDP formed quantified using ImageJ. Experiments were repeated three times with means and standard deviations shown. Statistical analysis was performed using a two-way ANOVA, followed by Dunnett’s multiple comparisons test (** p < 0.005, * p < 0.05). B) The RNA helicase activity of 0.5 μM CshA with and without RelSau was determined using a Cy3-labelled double stranded RNA oligomer. Reactions were incubated at 25°C for up to 10 min before analysis on a native page gel. Experiments were repeated four times with means and standard deviations shown. Statistical analysis was performed using a two-way ANOVA, followed by Sidak’s multiple comparisons test (* p < 0.05, **** p < 0.0001). C & D) rRNA extracted from LAC*. Strains were grown at 25°C until an OD600 of 0.4. Cells were split and half exposed to 60 μg/ml mupirocin for 1 h (plus), which at 25°C corresponds to one doubling. RT-PCR of total RNA using the probes as indicated in Fig 4C that bind internally to the 16S (C) or to the unprocessed leader and trailer sequences (D) of the 17S rRNA. rRNA values are expressed as mean relative expression +/- SD normalised to the expression of rho RNA as an internal control. For comparisons between two groups (C—LAC* minus v plus) a two-tailed, unpaired Student’s t-test was performed (**** p < 0.0001). For comparisons between more than two groups (D) a two-way ANOVA, followed by Sidak’s multiple comparisons test was performed (** p < 0.005). Cycle threshold values were determined for 3 biological repeats in duplicate (N = 3).