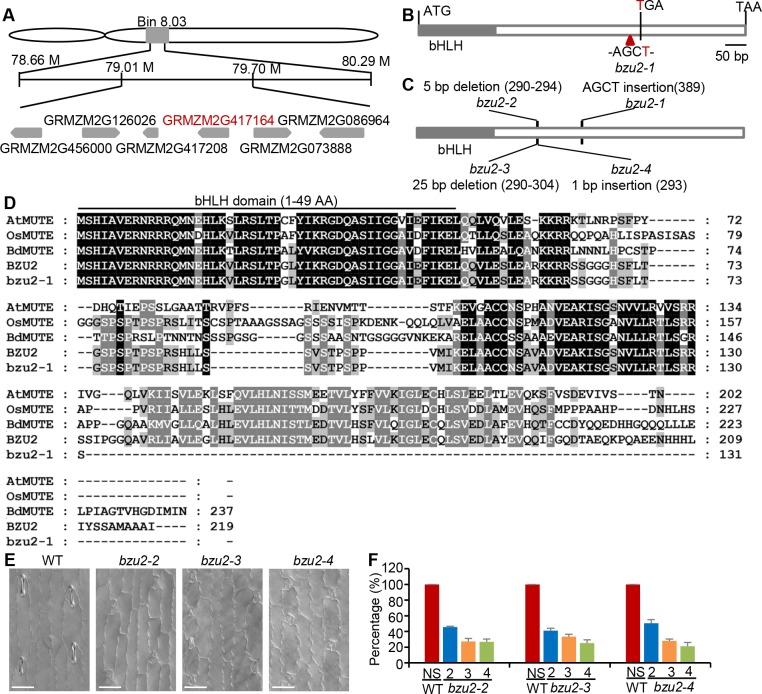
Fig 4. A mutation in BZU2 is responsible for stomatal defects in maize.
(A) BZU2 was mapped to bin 8.03 of chromosome VIII, between the 79M15 and 79M45 SSR markers, a 0.69 Mb region containing fifteen genes (the key gene is labeled in red). (B) The bzu2-1 mutation was generated by a 4 nucleotide AGCT insertion at position + 390 of GRMZM2G417164, which generated a stop codon within the reading frame. (C) Structure of bzu2-1 and the CRISPR/Cas9-induced mutations (bzu2-2, bzu2-3 and bzu2-4). bzu2-2 contains a 5 bp deletion downstream of the PAM site; bzu2-3 contains a 25 bp deletion downstream of the PAM site; bzu2-4 contains a 1 bp insertion downstream of the PAM site. (D) Alignment of BZU2 (GRMZM2G417164) (in wild-type and bzu2-1), AtMUTE (At3g06120), OsMUTE (LOC_Os05g51820) and BdMUTE (Bradi1g18400). (E) DIC images show similar aberrant GCs phenotypes, as well as a lack of SCs in bzu2-2, bzu2-3 and bzu2-4 mutants in the first and second leaves. Scale bars, 50 μm. (F) Percentage of stomatal complexes showing irregular GMC divisions in CRISPR/Cas9-induced mutations (bzu2-2, bzu2-3, and bzu2-4) in the T0 generation, based on examination of the first leaves of three 8-day-old wild-type and CRISPR/Cas9 mutants. In wild-type plants, all stomata are normal (NS, 100%). No normal stomata (NS, 0%) were observed in bzu2-2 (173 stomatal complexes examined), bzu2-3 (168 stomatal complexes), and bzu2-4 (150 stomatal complexes). Of 493 stomata surveyed, 48% comprised 2 cells, 28% comprised 3 cells, and 24% comprised 4 cells. Abbreviations: AA: amino acid, Normal stomata (NS), two-celled (2), three-celled (3), four-celled (4).