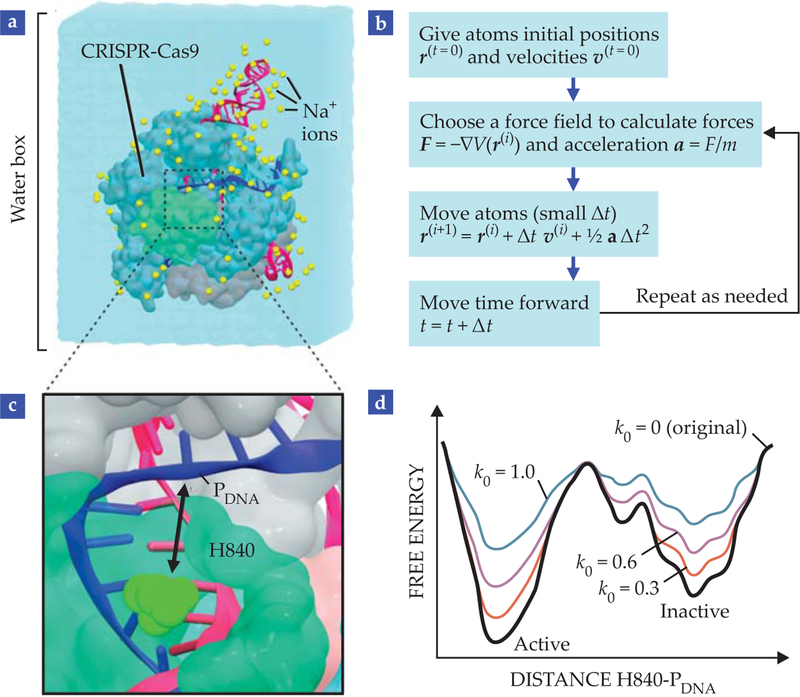
FIGURE 3. MOLECULAR DYNAMICS (MD) SIMULATIONS.
produce a temporal trajectory of (a) CRISPR-Cas9 embedded in a water box that contains sodium ions at physiological concentration. (b) In a nutshell, MD consists of giving atoms initial positions and velocities, choosing a set of functions and parameters to describe the forces acting on each atom, and advancing time using Newton’s equations of motion. The resulting trajectory of atomic coordinates can be used to track the system’s properties over time. (c) Gaussian accelerated molecular dynamics6 (GaMD) describes the movement of histidine H840 (the catalytic residue in HNH) to the cleavage site in the target strand (PDNA) for catalysis.10 (d) In GaMD, quadratic functions modify the original potential energy of the system in order to overcome the barrier between active and inactive states. The extent of acceleration is controlled by parameters of the Gaussian function. The greater the value of k0, the greater the acceleration and the easier the system overcomes the barrier between states.