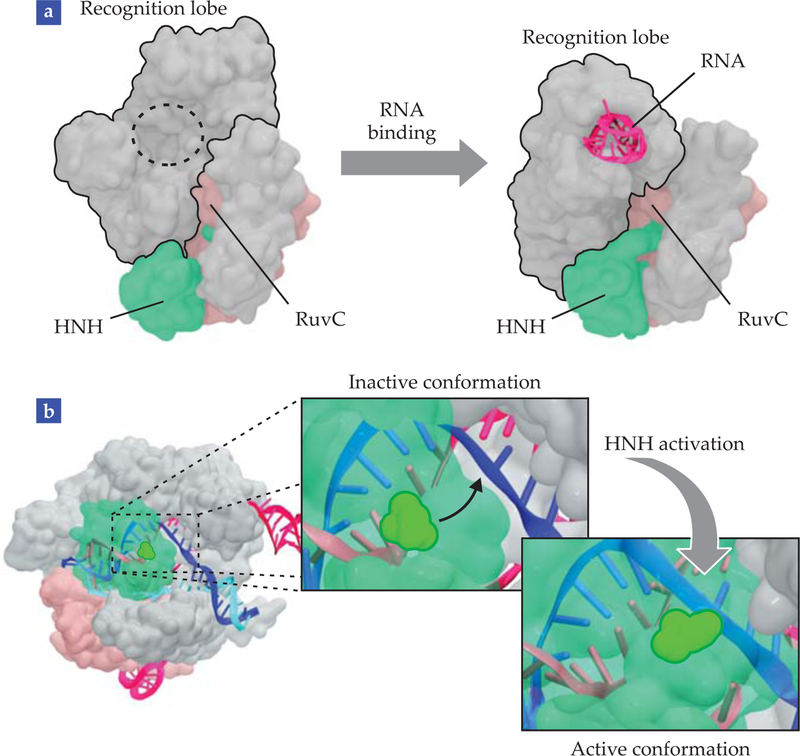
FIGURE 4. MOLECULAR MECHANISMS OF CRISPR-CAS9,
revealed by molecular dynamics simulations. (a) Rearrangements made to the recognition lobe reshape the molecular architecture of unbound Cas9 into its RNA-bound conformation. (b) The movement of HNH into its final catalytic state prepares it to cleave the DNA. The conformational change, shown as insets, brings the HNH catalytic residue close to the cleavage site in the DNA complementary strand. In all structures, Cas9 nuclease domains HNH and RuvC are green and pink, respectively. RNA is magenta; DNA is dark blue (complementary strand) and light blue (noncomplementary strand).