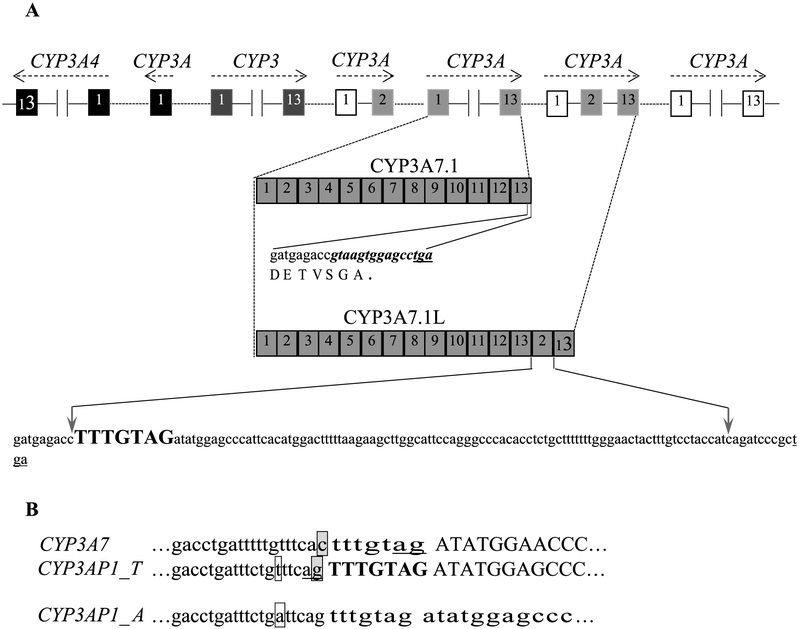
Fig. 1.
Location of CYP3A7 and its alternatively spliced variants. A: organization of CYP3A locus, sequences of the 3′ ends of CYP3A7 and its variant CYP3A7-3AP1. The canonical CYP3A7 last 15 bp (bold and italic) are replaced by the exons 2 and 13 of pseudogene CYP3AP1, generating a longer mRNA species CYP3A7-3AP1 with extra shift-mutated sequence. B: Intron 1-exon 2 boundaries in CYP3A7 and CYP3AP1. Introns and exons are shown in lowercase and in uppercase, respectively. The AG splice site of canonical CYP3A7 exon 2 is located in the 3′-end of the heptanucleotide spacer (tttgtag)(underlined). The 5′-flanking nucleotide of the heptanucleotide spacer (bold and uppercase) is a G (boxed and shaded) in CYP3AP1 rather than a C (boxed and shaded) in CYP3A7. This nucleotide variation shifts the AG acceptor splice site to an alternative site in CYP3AP1. The polymorphism (T > A, boxed) in CYP3AP1 prevents the pseudogene splicing.