Figure 6.
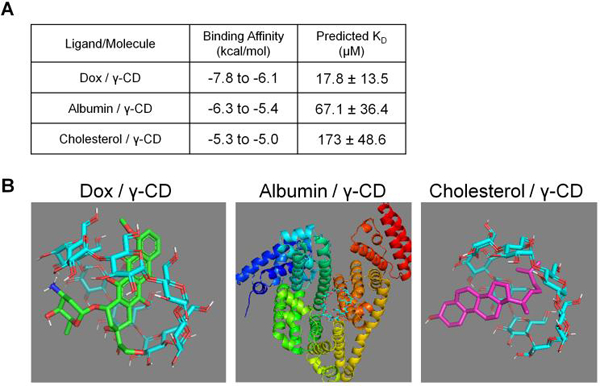
(A) PyRx binding simulation results of Dox, albumin, and cholesterol each individually binding with γ-CD. (B) Molecular models representing the highest affinity conformations for each simulated ligand/molecule pair in PyRx with Vina Autodock software. Note the scale is different and biological assemblies were simulated for Albumin / γ-CD.
