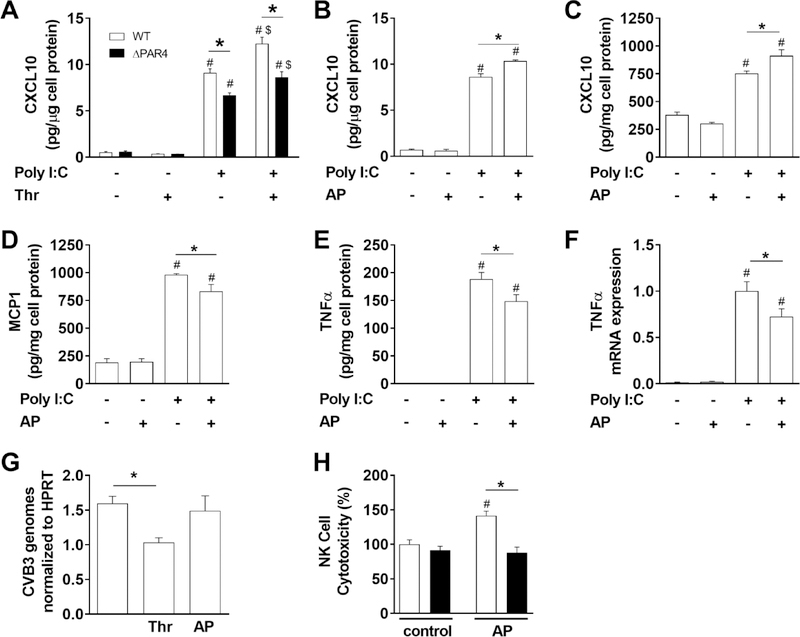
Figure 1. PAR4 activation modulates innate immune responses in vitro.
A) Levels of CXCL10 in cell culture media of bone marrow derived macrophages from WT (white bars) and PAR4−/− (ΔPAR4, black bars) mice in response to the dsRNA mimetic poly I:C (25 μg/mL) and/or thrombin (Thr, 5 nM) were analyzed after 7 hours stimulation. WT BMM (B) or WT murine embryonic fibroblasts (C) were stimulated with poly I:C (25 μg/mL) and/or PAR4 activating peptide (AP, 150 μM) for 7 hours and the CXCL10 protein levels analyzed in cell culture media by ELISA. WT MEF (D, E) were stimulated for 24 hours with poly I:C (25 μg/mL) and/or PAR4 AP (150 μM) and the MCP1 and TNFα protein levels were analyzed in the cell culture media by ELISA. F) TNFα mRNA expression was analyzed by real-time PCR in WT murine CM 4 hours after stimulation with poly I:C (25 μg/mL) and/or PAR4 AP (150 μM). Results are shown relative to poly I:C stimulation which was set to 1. G) CVB3 genomes were analyzed in WT murine CM by RT-PCR after infection with CVB3 (multiplicity of infection 1) and additional stimulation with thrombin (Thr, 5 nM) or PAR4 AP (150 μM) for 6 hours. Genome levels were normalized to HPRT mRNA expression. H) NK cell cytotoxicity against YAC-1 cells of splenocytes isolated from WT (white bars) and ΔPAR4 (black bars) mice after 24 hours stimulation with or without PAR4 AP (150 μM). Data (mean ± SEM; N>4) were analyzed by 2-way (A, H) and 1-way (B-G) ANOVA. Statistical significance is shown as * P<0.05, # P<0.05 versus unstimulated control within the same genotype or unstimulated control, and $ P<0.05 versus poly I:C alone.