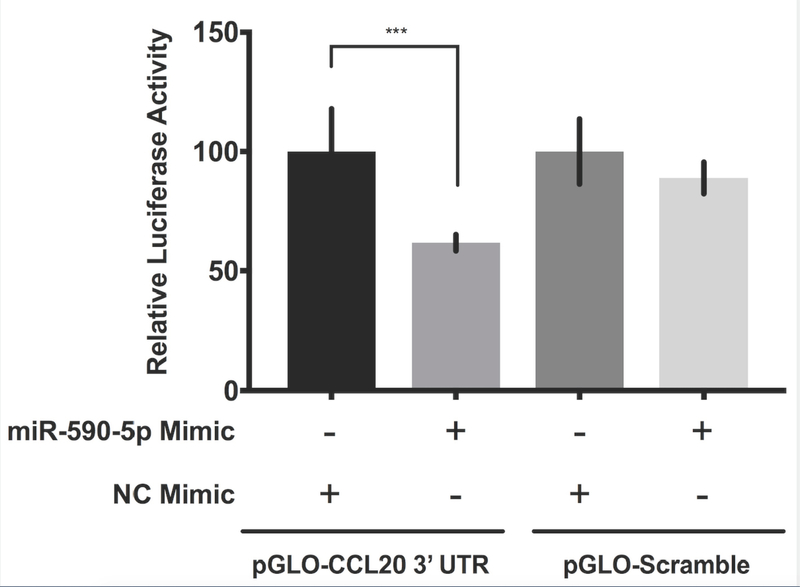
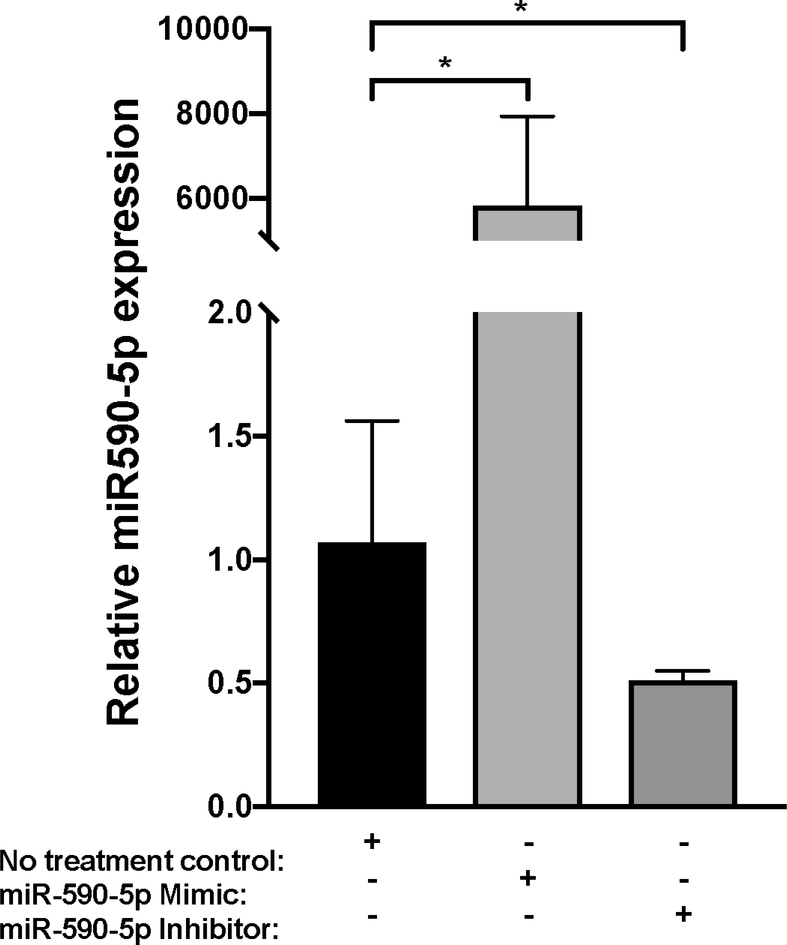
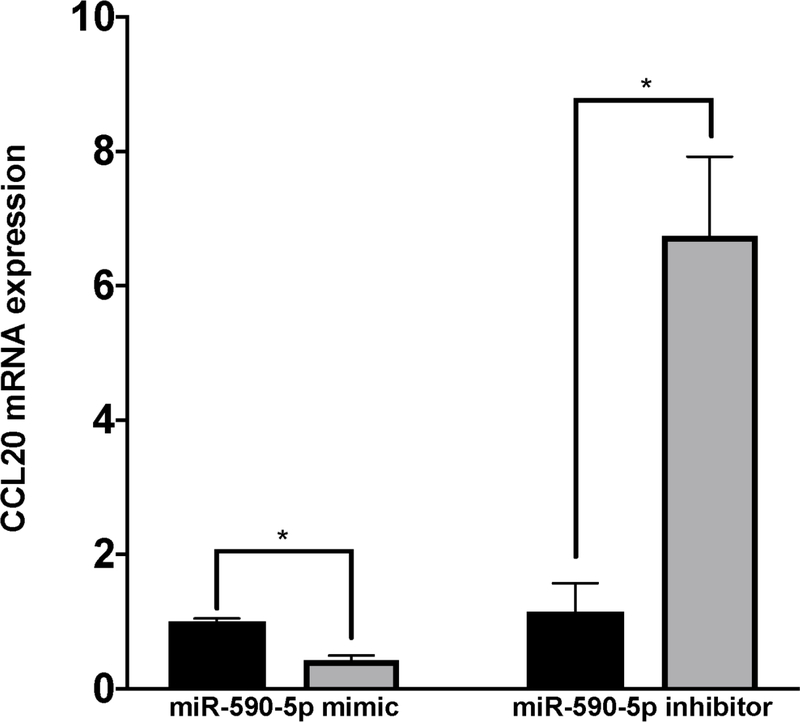
Figure 4. miR-590-5p functionally interacts with CCL20 to regulate its expression.
A) The miR-590-5p target site was predicted to align to positions 261–267 in the CCL20 3’UTR (GenBank Accession NM_004591). Vertical lines indicate paired alignment between the miRNA seed region and CCL20 3’UTR sequence. B) Approximately 2 × 105 human embryonic kidney 293 (HEK293) cells were co-transfected with the pGLO-CCL20 3’UTR vector or scrambled control vector (pGLO-Scramble) and miR-590-5p mimic or scrambled sequence control miRNA (negative control [NC] mimic), and luciferase activity was measured 48 hours post-transfection using the Dual-Glo Luciferase Assay System (Promega). The ratio of firefly luciferase activity to renilla luciferase activity was determined for each sample and the firefly/renilla relative luminometer units (RLU) in HEK293 cells co-transfected with miR-590-5p mimic and the pGLO-CCL20 3’UTR vector were compared with those from cells co-transfected with pGLO-Scramble and NC mimic. The Mann-Whitney U test was used to assess differences between conditions; differences between the negative controls were not statistically significant. Activated LX-2 cells were transfected with 75 nM mirVana miRNA-590-5p mimic, 50 nM anti-miR-590-5p inhibitor, or 50 nM scrambled sequence control. After 48 hours cells were harvested and total RNA was extracted.
The TaqMan RNA-to-Ct 1-Step kit (Thermo Fisher Scientific) and TaqMan commercial primers were used to measure C) miR-590-5p and D) CCL20 transcript levels in the presence of miR-590-5p mimic and miR-590-5p siRNA. Cycle threshold levels were generated using QuantStudio Real-Time PCR Software 1.0 and mRNA data were normalized using GAPDH. Black bars represent the scrambled miRNA negative control, while the gray bars represent treatment conditions. All experiments were performed in triplicate. Data are means ±SD. *P<0.05; **P<0.001; ***P<0.0001.