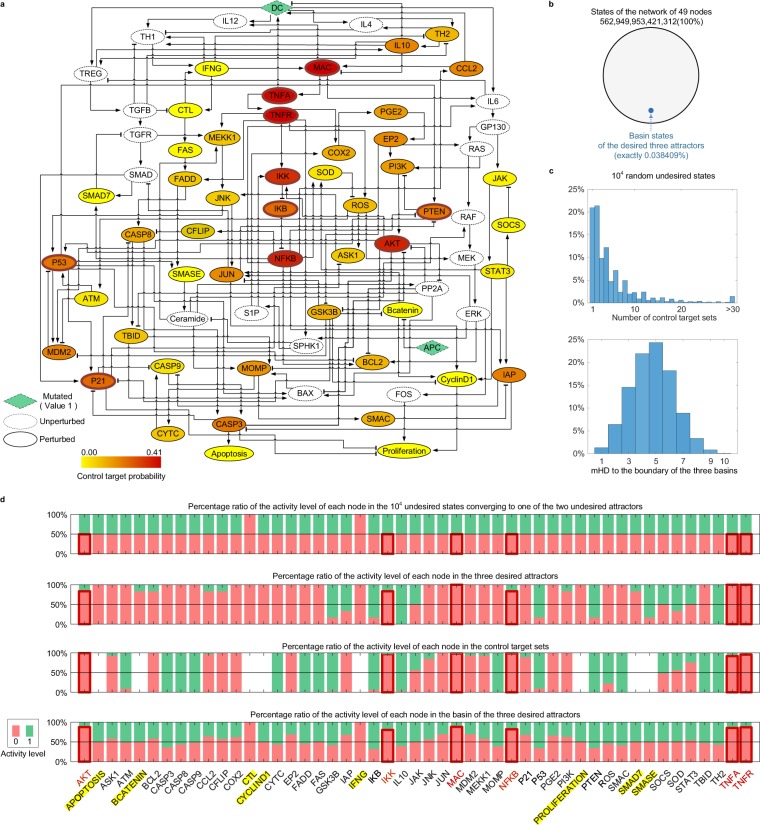
Figure 3.
Application of BRC to the CACC49 network having a relatively small basin of the desired attractor. (a) CACC49 Boolean network with the probability of each node being a control target. Arrows and bar-headed lines represent activation and inhibition, respectively. Nodes DC and APC have mutated ON state (value 1), leading to fixed state values of the white nodes (dotted circles). Nodes with solid circles denote the candidate control targets. 10,000,000 randomly selected initial states are used to find out undesired attractors with constantly activated Proliferation (value 1) and desired attractors with constantly inactivated Proliferation (value 0), which are given in Supplementary Data 2. The darker red color of a node denotes a higher probability of being contained in control target sets given 10,000 randomly selected undesired states. (b) Sufficiently many basin states in relatively small size of a desired basin. Using the 10,000,000 randomly selected initial states, three desired attractors with constantly inactivated Proliferation are found (Supplementary Data 2). The size of desired basin states is exactly 0.038409% (21,622,344,760,959 states). (c) Distributions of the numbers of control target sets and mHD. Symbol “>30” in the top panel indicates that the number of control target sets is greater than 30. The average number of control target sets is 5.2476 (standard deviation 5.7703) and the average mHD is 4.7426 (standard deviation 1.5885). (d) Nodes enriched in control target sets. The activity levels range from 0 (inhibition) to 1 (activation). The first, second, third and last panels denote the activity level percentages of each node in 10,000 randomly selected undesired states, the desired attractors’ states, the control target sets and the desired basin states, respectively. The heights of boxes with red outlines in the four panels denote the activity level percentages of MAC, TNFA, TNFR, NFKB, AKT and IKK (darker red colors in a) being contained in control target sets, which are the top 6 control target nodes. The nodes marked with yellow backgrounds (APOPTOSIS, BCATENIN, CTL, CYCLIND1, IFNG, PROLIFERATION, SMAD7 and SMASE) indicate that they are not control target nodes (denoted by empties in the third panel).