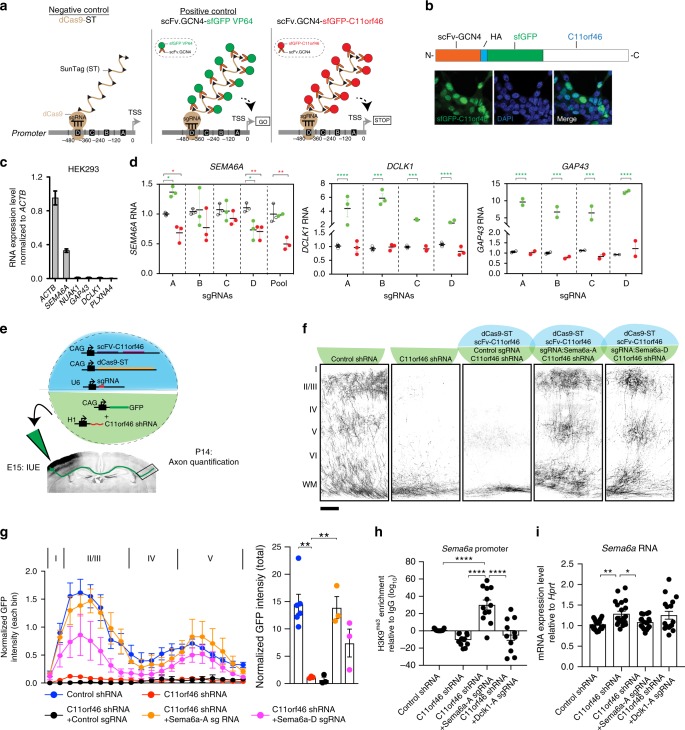
Fig. 5.
C11orf46- epigenomic editing of neurite-regulating genes rescues transcallosal dysconnectivity in C11orf46-deficient neurons. a Schematic representation showing epigenome editing using nuclease-deficient CRISPR/Cas9 SunTag (dCas9-ST) system. Single guide RNA (sgRNA) targeting neuronal gene promoters is introduced with dCas9-ST (10xGCN4) and scFv-sfGFP-C11orf46 (or −VP64 as a positive control) in HEK293 cells. b Overexpressed fGFP-C11orf46 is colocalized with DAPI signals in HEK293 cells. c Relative expression levels of neurite-regulating genes in HEK293 cells. d dCas9-ST mediated promoter loading with 10xC11orf46, and C11orf46R236H in comparison to 10xVP64 at SEMA6A in HEK293 cells. Four sgRNAs (a–d) for each gene were tested individually, and for SEMA6A also as a pool. Two-way ANOVA followed by Dunnett’s multiple comparisons test was performed where C11orf46- or VP64-epigenetic editing effects were tested. e Epigenomic editing of transcallosal neurons by IUE. Five transgenes were delivered simultaneously at E15 by IUE into the developing somatosensory cortex, followed by quantification of axonal arborization at P14. f, g Axonal arborization was disrupted by C11orf46 knockdown, and restored by dCas9-ST mediated recruitment of C11orf46 to Sema6a promoter (Sema6a-), using sgRNAs Sema6a-A, Sema6a-D, but not with non-targeting sgRNA. Scale bar, 100 µm. Layer distributions and total axonal density in the contralateral site of the electroporated brains are shown (g). F (4, 13) = 13.1934, P = 0.0002 was determined by one-way ANOVA with post hoc Bonferroni test. Three to six mice per condition. h H3K9me3 enrichment at Sema6a promoter in GFP + neurons at P0, four days after IUE. Epigenome editing by Sema6a-A sgRNA increases H3K9me3 levels at Sema6a promoter. Dclk1-A sgRNA did not affect H3K9me3 levels at Sema6a promoter. F (3,44) = 8.718, P = 0.0001 was determined by one-way ANOVA with post hoc Bonferroni test. i Sema6a mRNA level in GFP + neurons at P0. Increased Sema6a expression caused by C11orf46 knockdown were suppressed by C11orf46-epigenome editing using Sema6a-A sgRNA, but not by Dclk1-A sgRNA. F (3,72) = 4.746, P = 0.0045. *P < 0.05, **P < 0.01 determined by one-way ANOVA with post hoc Bonferroni test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Bar graphs indicate mean ± S.E.M.