Fig. 3.
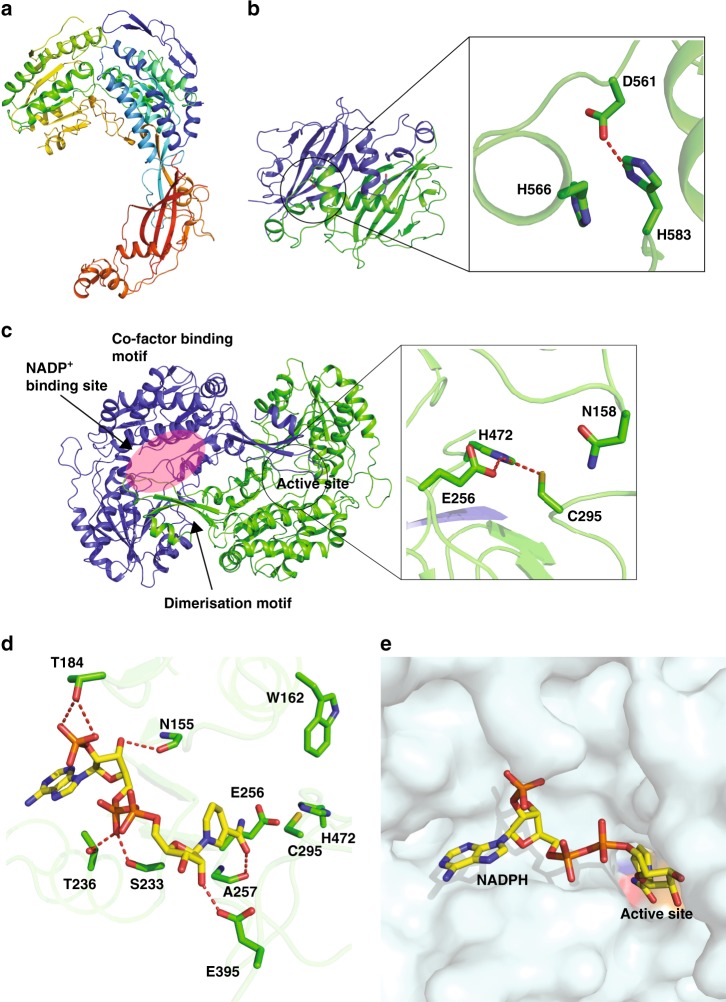
Enzymatic domains in PaaZ and the cofactor binding site. a The polypeptide of the PaaZ monomer color coded in rainbow with the N-terminus in blue and C-terminus in red. b The hydratase domain is made of five β-sheets with three additional helices and the monomers are colored in blue and green. The active site comprising D561 and H566 of one monomer is closer to the substrate-binding tunnel of the other monomer, and a hydrogen bond (3.3 Å) is observed between H583 and D561. c The dehydrogenase domain can be divided into three sub-domains: co-factor binding, catalytic and dimerization23. They are made of a mixture of α-helices and β-strands. The active site of the dehydrogenase domain consists of nucleophile cysteine C295 and E256. A histidine residue (H472) is in hydrogen bonding distance to E256 and ~3.55 Å to C295. The distance between E256 and C295 is 3.5 Å. The monomers of the dehydrogenase domain are colored in blue and green. d The interactions of NADPH observed in the structure. Numerous interactions with polar and charged residues of the enzyme stabilize the NADPH binding. e The NADPH binding groove and the active site residues are colored (C295 in orange, E256 in red and H472 in blue). The nicotinamide moiety points towards the active site cavity