Fig. 5.
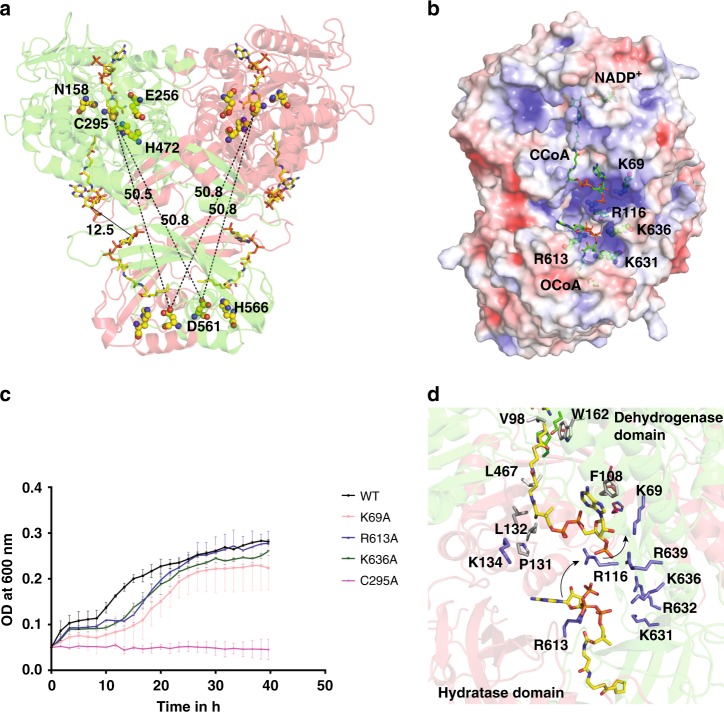
Substrate transfer pathway in PaaZ. a Cartoon representation of PaaZ dimer, monomers colored in red and green. The active site residues and ligands are shown in sphere and stick representation, respectively. The distance between the active site residues between the two catalytic centers measured from the same monomer or adjacent monomer is ~51 Å. The ligands OCoA and CCoA from the two structures are used as reference to show the relative positions of the substrates in each enzymatic domain. The distance between the substrates measured from C3 carbon is ~12.5 Å. Note that the OCoA ligand placed in the hydratase domain of the PaaZ CCoA structure is not an experimental structure. b Electrostatic (ABPS) potential of PaaZ dimer colored between −5 to 5 kT. The positive patches at this interface region indicate that the substrate from the hydratase domain might be attracted and guided into the tunnel of the dehydrogenase domain. Key positively charged residues are shown in sphere representation. c Growth curves of E. coli K12 ΔpaaZ transformed with plasmid encoding PaaZ variants—WT, K69A, R613A, K636A and the control C295A. All 3 alanine mutants of the positive charged residues showed slower growth compared to WT, when grown in a minimal media with PA as the sole carbon source. The mutant K69A was the slowest suggesting its role in transfer of substrate from hydratase domain to dehydrogenase domain. Error bars reflect the s.d. of three independent experiments (n = 3). The source data are provided as a Source Data File. d A close up view of the substrate transfer pathway in PaaZ, colored as in panel a. The possible mode of substrate transfer is shown with black arrow. Conserved residue R613 stabilizes the substrate in the hydratase domain and upon ring opening, the substrate could potentially interact with R116. The presence of other charged residues (K636, K631 and R632) are likely to stabilize or guide the substrate toward R116 and the dehydrogenase domain. Subsequently, the hydrophilic tail is stabilized by K69 and only in this step, is the hydrophobic head group likely to be flipped in to the tunnel of the dehydrogenase domain