Abstract
Reverse transcription quantitative PCR (RT-qPCR), a method of choice for quantification of gene expression changes, requires stably expressed reference genes for normalization of data. So far, no reference genes were established for the Alphaproteobacteria of the genus Ochrobactrum. Here, we determined reference genes for gene expression studies in O. quorumnocens A44. Strain A44 was cultured under 10 different conditions and the stability of expression of 11 candidate genes was evaluated using geNorm, NormFinder and BestKeeper. Most stably expressed genes were found to be rho, gyrB and rpoD. Our results can facilitate the choice of reference genes in the related Ochrobactrum strains. O. quorumnocens A44 is able to inactivate a broad spectrum of N-acyl homoserine lactones (AHLs) – the quorum sensing molecules of many Gram-negative bacteria. This activity is attributed to AiiO hydrolase, yet it remains unclear whether AHLs are the primary substrate of this enzyme. Using the established RT-qPCR setup, we found that the expression of the aiiO gene upon exposure to two AHLs, C6-HLS and 3OC12-HSL, does not change above the 1-fold significance threshold. The implications of this finding are discussed in the light of the role of quorum sensing-interfering enzymes in the host strains.
Subject terms: Bacterial transcription, Bacterial genes
Introduction
Reverse transcription quantitative PCR (RT-qPCR) is the most widely used approach for gene expression studies. However, despite being powerful, the method involves multiple steps and is sensitive to both flaws in experimental design and to technical variation.
One of the crucial decisions to make while performing a RT-qPCR experiment is the choice of stably expressed reference genes (RGs) for the normalization of data. In case of model or well-studied organisms, the selection of suitable RGs can be facilitated based on previous studies. For bacteria, literature provides a list of candidate RGs of potential general use due to their occurrence in multiple species. The most popular include rho, 23S rRNA, rpoD, gyrB1, recA1–3, 16S rRNA4–6, dnaK, rpoB2,7, groEL7, gyrA8. However, these candidates are not equally suitable for all species under all growth conditions and one should always experimentally verify the stability of a set of candidates when first-time applying RT-qPCR in the given (micro)organisms9,10. In 2009, a set of guideline was published called ‘Minimum Information for publication of Quantitative real-time PCR Experiments’ (MIQE)11. Its goal was to unify the quality of RT-qPCR results published in scientific journals, mainly concerning medical studies. In prokaryotic studies, with the exception of a few earlier works (i.a.1), the pursuit to meet the MIQE standards intensified only recently10,12,13. Still, in many publications concerning RT-qPCR in bacteria, the expression of the studied gene(s) is arbitrary normalized to certain RGs without validation of their expression stability or even without a clear explanation for the choice.
One of the bacterial genera for which no data has been available on the suitable RGs is Ochrobactrum spp. These ubiquitous Alphaproteobacteria, related to Brucella, Agrobacterium and Rhizobium14–17, attract scientific attention as opportunistic human pathogens14,17 but also as beneficial members of plant and nematode microflora and bioremediation agents18–29. In this study, we focused on the Ochrobactrum quorumnocens strain A4430, an isolate from the rhizosphere of potato formerly known as Ochrobactrum sp. A4431. As a result of the activity of AiiO hydrolase, A44 inactivates a broad spectrum of bacterial signaling molecules from the group of N-acyl homoserine lactones (AHLs)32. Many Gram-negative bacteria secrete and autodetect AHLs, responding at transcriptional level when a threshold concentration is reached. This regulatory mechanism, termed quorum sensing (QS), enables populations of single-celled organisms to perform coordinated actions such as bioluminescence, biofilm formation or production of virulence factors33,34. The functioning of QS can be disturbed by AHL-cleaving microorganisms like O. quorumnocens A44, resulting in the loss of the QS-dependent phenotype35–37. The latter phenomenon is known as the quorum quenching (QQ). For example, A44, when co-inoculated on plant tissue with the plant pathogenic bacterium Pectobacterium parmentieri SCC3193, attenuates the QS-governed virulence of this pathogen31,38.
To date, numerous AHL-inactivating enzymes have been described39,40. Discovery of many of them, including AiiO, resulted from screening studies designed for the selection of new QS-disrupting agents. It is unclear whether the cleavage of AHLs in some bacterial species is the primary function of the involved proteins41. For some of these enzymes, additional, unrelated functions were identified in the respective hosts, indicating that AHL cleavage may be the effect of catalytic promiscuity (reviewed in40,41). For example, BlcC (AttM) from Agrobacterium tumefaciens was found to be involved in the metabolism of gamma-aminobutyrate42, and PvdQ from Pseudomonas aeruginosa is necessary for the maturation of the pyoverdine siderophore43. Even if the function of certain enzymes is assumed to be inactivation of AHLs, the predicted biological purpose of this activity is not uniform among the QS-interfering strains. Depending on whether the strain produces its own AHL or not, the predicted functions include, but are not limited to, self-regulation of own QS, using AHLs as a source of nutrients and providing an advantage in the environment over the AHL-producing competitors (reviewed in40). The role of the AHL-cleaving enzyme AiiO in the metabolism and environmental fitness of O. quorumnocens A44 remains unclear.
In this study, we selected stably expressed RGs for RT-qPCR analyses in O. quorumnocens A44. Next, we applied RT-qPCR to measure changes in the expression levels of aiiO in A44 in response to AHLs and factors such as growth phase, growth temperature, the medium pH the addition of potato root extract. The study is a part of our investigation on the role of AiiO in the metabolism and fitness of O. quorumnocens A44.
Results
Selection of reference genes (RGs)
A gfp-tagged variant of O. quorumnocens A44 was used throughout the study (hereafter referred to as A44)44. Eleven candidate genes were explored as potential RGs. Nine candidate genes (16S rRNA, 23S rRNA, dnaK, groEL, gyrB, recA, rho, rpoB and rpoD) were chosen based on previous reports on RGs used in bacteria1–7 (Supplementary Table S1). The remaining two candidates were: gfp, constitutively expressed by the tagged strain from an artificially introduced vector pPROBE-GTkan45, and CES85_4722, an A44 gene encoding a hypothetical protein with no data concerning its stability (a blind control). The gfp was explored as an RG candidate due to its rare occurrence in the environment, a feature that would be valuable for potential studies on complex samples. Primers targeting the aforementioned genes in A44 were designed and specificity in amplification of fragments of expected sizes (139–148 bp) was confirmed by gel electrophoresis and a melt curve analysis (Supplementary Fig. S1AB). In parallel, the performance of the primers targeting the aiiO gene was verified.
Expression stability of candidate RGs
A pilot assay was conducted to establish the expression stability of 11 candidate RGs under 10 different conditions (Table 1).
Table 1.
Conditions for the growth of A44 strain applied for the RT-qPCR experiment.
| Basal mediuma | Supplements | pHb | Growth temp. (°C) | Growth phasec |
|---|---|---|---|---|
| LB (L) | — | 5.5 | 28 | early stat. |
| LB (L)4 | — | 7 | 28 | early stat. |
| LB (L) | — | 7 | 28 | middle log. |
| LB agar (S) | — | 7 | 28 | O/N |
| LB agar (S) | — | 7 | 20 | O/N |
| LB agar (S) | — | 7 | 37 | O/N |
| LB agar (S) | co-culture with P. parmentieri SCC3193 | 7 | 28 | O/N |
| M63 0.4% glucose (L) | 7 | 28 | O/N | |
| M63 0.4% glucose (L) | 50 μM C6-HSLd | 7 | 28 | O/N |
| M63 0.4% glucose (L) | 50 μM 3OC12-HSLe | 7 | 28 | O/N |
| M63 0.4% glucose (L) | 25% potato roots extractf | 7 | 28 | O/N |
1(L) and (S) stand for liquid and solid medium, respectively.
aThe medium was buffered only in case of pH 5.5. In other cases, 7 is the initial pH of the medium.
bIf provided, growth phase was determined based on the growth curve of A44 GFP in LB at 28 °C. O/N – stands for an overnight culture (16–20 h). early stat. – stands for early stationary growth phase.
cCulture condition not included in the geNorm pilot experiment (initial ranking of RG candidates).
dC6-HSL – N-hexanoyl homoserine lactone.
e3OC12-HSL – N-3-oxododecanoyl homoserine lactone.
fThe medium contained water extract from the roots of soil-grown, 3-week-old potato plants (final concentration 25%).
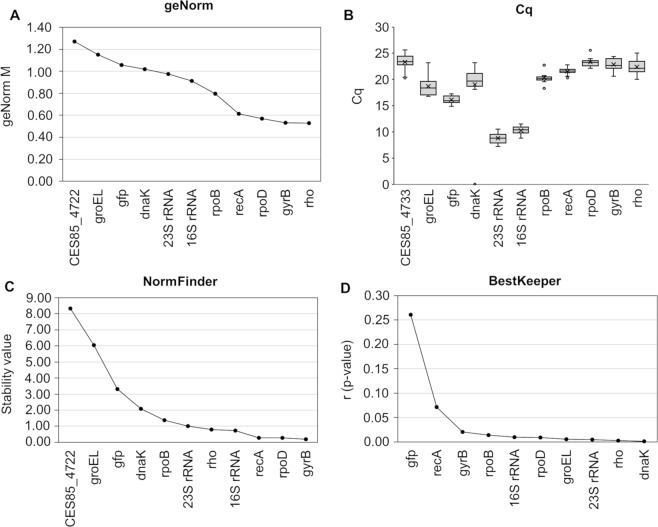
According to geNorm46, the most stably expressed candidate RGs, characterized by the lowest geNorm M values (≤0.6), were found to be rho, gyrB and rpoD, followed by recA (Fig. 1A). The commonly applied RG encoding 16S rRNA was considerably less stable (M = 0.913). The heterologously expressed gfp gene, with M = 1.058, was also not among the top-ranking candidates. The mean threshold quantification cycle (Cq) obtained for the 16S rRNA was 10.26 and 8.79 for the 23S rRNA. Therefore, the Cq for these RG candidates was 6–13 and 7–15 cycles lower, respectively, than that of the other candidates, reflecting the high abundance of the rRNA species in the cells (Fig. 1B).
Figure 1.
Expression stability of candidate RGs in O. quorumnocens A44 obtained using the respective algorithms: geNorm (A), NormFinder (C) and BestKeeper (D). Panel B shows Cq values for the investigated genes. The stability of expression of the RGs candidates was investigated under 10 different culture conditions. As BestKeeper enables simultaneous analysis of up to 10 genes, the blind control CES85_4722 (the eleventh gene), being the least stable gene according to geNorm and NormFinder, was excluded from the set for this tool.
The results obtained with geNorm were compared with RG ranks obtained with two other popular algorithms: NormFinder47 and BestKeeper48. The 4 genes selected by geNorm were also among the top 5 ranked by NormFinder, accompanied by the 16S rRNA gene (Fig. 1C). For three best-ranking genes, gyrB, rpoD and recA, NormFinder assigned stability values of 0.19, 0.28 and 0.28, respectively. BestKeeper ranked all analyzed genes except for gfp and recA as well-performing, that is with a statistically significant (α = 0.05) correlation between the correlation coefficients of individual genes with the BestKeeper index calculated for all genes in the analysis (Fig. 1D). A juxtaposition of the results from the three algorithms ranked rho, rpoD and gyrB as the most stable genes (Supplementary Table S2).
Apart from ranking genes by their stability, the geNorm tool can also suggest the optimal number of RGs to be included in the normalization factor (NF)46. In this study, geNorm tool suggested using three RGs (V value < 0.15 threshold) (Fig. 2). Therefore, the complete set of samples (11 conditions, 2–3 biological replicates) for the analysis of the expression of aiiO (gene of interest) was analyzed by targeting aiiO alongside the three most stable RG candidates: rho, rpoD and gyrB. Next, the stability of expression of the applied RGs was validated on the complete dataset. geNorm analysis indicated the highest stability for rho (M = 0.441, CV = 0.187), followed by gyrB (M = 0.453, CV = 0.185) and rpoD (M = 0.5, CV = 0.221).
Figure 2.
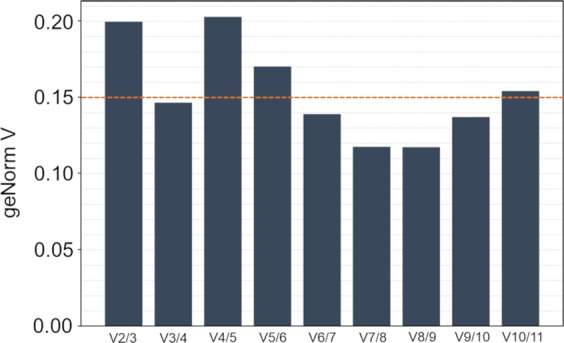
Optimal number of reference genes for normalization of RT-qPCR data suggested by geNorm. The tool calculates the pair-wise variation Vn/Vn + 1, where n is the number of reference genes used for normalization. According to Vandesompele and co-workers46, V value below 0.15 (dotted line) signifies that inclusion of an additional gene (n + 1) to the normalization factor yields no benefit.
PCR efficiency
PCR efficiency is a parameter included in the final calculation of the relative abundance of the investigated targets. With the primers designed in this study, PCR efficiency for gyrB and rho were 95.9% and 93.5%, respectively, and similar to the efficiency for the gene of interest aiiO (94.9%). The efficiency of PCR for rpoD was slightly lower (89%) (Table 2; Supplementary Fig. S2).
Table 2.
Validation of RT-qPCR assay parameters for primers targeting aiiO, gyrB, rho and rpoD.
| Parametera | aiiO b | gyrB | rho | rpoD |
|---|---|---|---|---|
| Efficiency (%) | 94.9 | 95.9 | 93.5 | 89 |
| R2 | 0.999 | 1 | 1 | 0.999 |
| Slope | −3.451 | −3.425 | −3.487 | −3.618 |
aEfficiency – calculated primer efficiency; R2 – correlation between log Cq and Cq in the standard curve; Slope – slope of the standard curve.
bPrimers used for PCR amplification: aiiO: F_aiiO_OP7/R_aiiO_OP7; gyrB: F_ gyrB_OP7/R_ gyrB_OP7; rho: F_ rho_OP7/R_ rho_OP7 (Table S1).
Comparison of the performance of the selected normalization factors (NFs)
GeNorm analysis recommended applying 3 RGs for data normalization in the developed RT-qPCR assay. To determine whether the change of RGs included in the NF may influence the interpretation of the expression data for our gene of interest (aiiO), we compared the consistency of results obtained using 3 different NFs: NF1 (rho, the most stably expressed RG based on geNorm), NF2 (rho, gyrB) and NF3 (rho, gyrB and rpoD). For the comparison of the single-gene NF1 and the 2-gene NF2, the results were significantly different (p < 0.05) under 3 out of 11 culture conditions tested (LB agar 28 °C, LB early stationary, LB middle exponential) (Fig. 3). For the comparison of the single-gene NF1 and the 3-gene NF3, the results were significantly different in 4 conditions (LB agar 28 °C, LB agar 20 °C, LB early stationary, LB middle exponential). On the contrary, the difference between applying the 2-RG NF2 and the 3-gene NF3 was significant only under a single culture condition (M63 + 3OC12-HSL) (Fig. 3).
Figure 3.
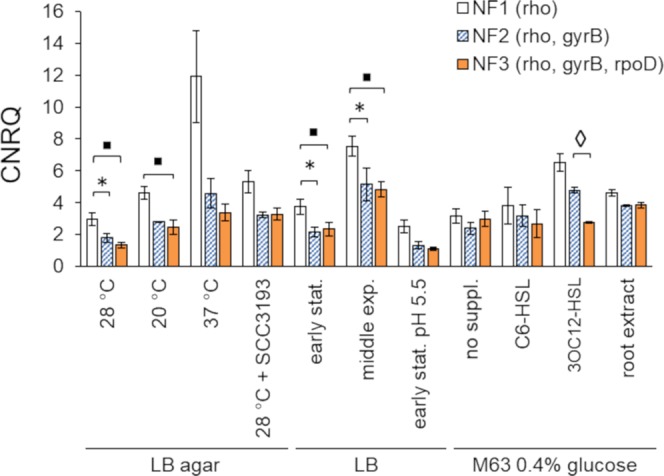
Influence of the type of the applied normalization factor (NF) on the relative expression of aiiO under different culture conditions. Statistically significant differences between groups (p < 0.05) are marked with: asterisk (NF1 vs NF2), squares (NF1 vs NF3) and diamonds (NF2 vs NF3). CNRQ – calibrated normalized relative quantity.
Changes in the expression level of aiiO in response to AHLs and other culture conditions
Expression of aiiO was investigated in three types of media: lysogeny broth (LB) with peptone as the main energy source, LB solidified with agar and a defined mineral medium M63 supplemented with 0.4% glucose. Based on these media, a set of 11 conditions was established, 10 of which were included in the study on the stability of RGs (Table 1), in order to test whether the expression of aiiO in A44 is affected by factors such as: (i) the growth temperature, (ii) growth phase, (iii) addition of potato root extract and finally (iv) the presence of chosen AHLs. Two of the AHL types, N-3-oxo-octanoyl homoserine lactone (3OC8-HSL) and N-3-oxohexanoyl homoserine lactone (3OC6-HSL), were delivered as compounds secreted by the co-cultured cells of P. parmentieri SCC319338. N-hexanoyl homoserine lactone (C6-HSL) and N-3-oxododecanoyl homoserine lactone (3OC12-HSL) were delivered as commercially synthesized pure compounds in 50 µM concentration. All AHLs included in the assay are known to be degraded by A44. To prevent complete depletion of AHLs prior sampling of cells for RNA extraction, the sampling was performed 90 min post addition of AHLs. The time point was chosen based on a previous experiment in which C6-HSL, supplemented to the culture in a 5-time lower concentration (10 µM), could still be detected in the supernatant of culture of A44 at 40 min (strong signal) and 100 min (weak signal) post addition31.
The expression data for aiiO was normalized with respect to NF3 and the resulting CNRQs (Supplementary Table S3) were grouped by the type of basal medium (LB, LB agar, M63 0.4% glucose). For each of the 3 types of medium, one sample was established as a reference (‘control’). The control samples were derived from cells grown in a respective medium without any additives and at 28 °C. When compared to the control samples, expression of aiiO was not altered above 1.5-fold in any of the tested conditions (Fig. 4). The >1-fold threshold was breached for LB culture in acidic pH 5.5, with respect to non-buffered LB medium of initial pH 7 (downregulation, −1.090 ± 0,116, p < 0.0005). The 1-fold threshold was also exceeded for LB agar cultures grown at 37 °C (upregulation, 1.320 ± 0.228) and at 28 °C in the presence of SCC3193 (1.286 ± 0.159), with respect to cultures grown separately at 28 °C, and in the middle exponential phase cells in LB with respect to early stationary phase cells in the same medium (1.023 ± 0,147). However, the latter three changes were not statistically significant (p > 0.05). Difference between M63% glucose and M63% glucose supplemented with 3OC12-HSL, although significant from a statistical standpoint, was below the 1-fold change cut-off (−0.115 ± 0.026, p < 0.05) (Fig. 4, Supplementary Table S4). In general, the expression of aiiO was not considerably induced under any of the conditions tested. In contrast, the expression of groEL, one of the least stable RG candidates in this study, was nearly 6-fold higher in samples from the mid-exponential culture in LB than in samples from LB agar at 37 °C, again with no significant induction observed for aiiO (Supplementary Fig. S3).
Figure 4.
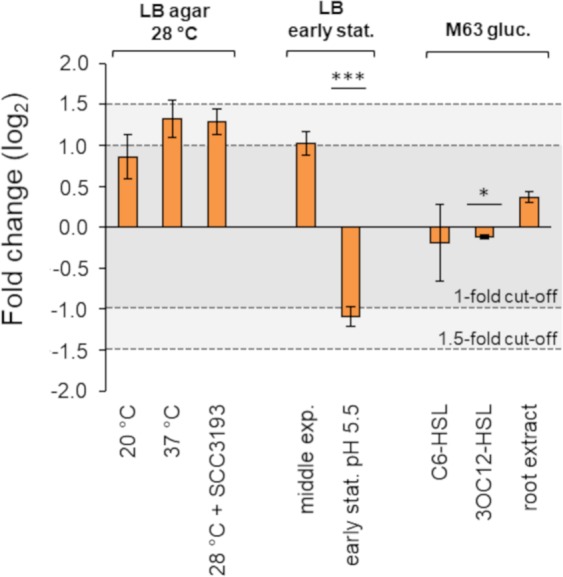
Fold change in relative expression of aiiO under different culture conditions. Relative expression (CNRQ) of aiiO in all samples was calculated with respect to normalization NF3 comprising rho, gyrB and rpoD. The fold change (log2) was calculated independently for each type of basal medium (LB agar, LB and M63 0.4% glucose) where the median CNRQ of one of the samples was treated as a reference (‘control’). If not indicated otherwise, the cells were grown at 28 °C. SCC3193 – growth in the presence of AHLs secreted by P. parmentieri SCC3193; no suppl. – without supplementation; early stat. – early stationary growth phase; middle exp. – middle exponential growth phase; C6-HSL, 3OC12-HSL – aiiO expression 90′ after the addition of 50 μM·mL−1 of the respective AHL; root extract – growth in the presence of 25% water-based extract obtained from potato roots. *denotes p < 0.05 ***denotes p < 0.0005 calculated using BootstRatio.
Discussion
The QQ activity of O. quorumnocens A44 is attributed to the α/β hydrolase protein AiiO32. However, little is known concerning the role of this enzyme in the metabolism and environmental fitness of this strain.
Here, we developed a RT-qPCR assay to study the expression of aiiO gene in O. quorumnocens A44. A key consideration in RT-qPCR is the choice of stably expressed reference genes (RGs), also known as internal controls. To identify RGs suitable for RT-qPCR analyses in O. quorumnocens A44, we tested the expression stability of 11 candidate genes, 9 of which were previously reported as RGs in other bacterial species (i.a.1–7). Despite it is known that RGs suitable for one microorganism often do not show sufficient stability in others (RGs are not universal)9, and that the incorporation of inadequate genes to the normalization factor (NF) can significantly alter the results12, the experimental validation of the stability of RGs tends to be neglected in gene expression studies in prokaryotes.
Ideally, the expression stability of a potential RG should be validated in all physiological or experimental conditions intended for the study of the gene of interest13. Here, we ranked the stability of expression of 11 candidate RGs in a pilot assay including 10 different conditions, on samples originating from a single experiment, and then verified the performance of best RGs in the final dataset (11 culture conditions, all biological replicates). The strategy for applying a pilot assay to choose the most promising RG candidates is in line with practical recommendations for the implementations of MIQE49. When human clinical samples are analyzed, a minimum of 10 samples from different patients can be used to rank RG candidates and then the best scoring RGs, usually 2–5, are targeted alongside the gene(s) of interest for all samples/patients under investigation50. Although a sample set derived from a bacterium cultured in 10 different conditions is not equivalent to a sample set from 10 human individuals, the tools developed for the evaluation of gene expression stability usually require an input of this approximate size. Therefore, this approach is also used for prokaryotic studies10.
The expression stability of the candidate RGs was evaluated using geNorm46 and two other popular algorithms, NormFinder47 and BestKeeper48, each relying on a different statistical approach9,51. According to geNorm, the most stable RG candidates were rho, gyrB and rpoD. These genes were also among the five top ranking genes according to NormFinder. The ranking of gene stability suggested by Bestkeeper was different from that suggested by the other two algorithms. This method also yielded the smallest resolution, assigning good performance to the majority of tested genes. Interestingly, recA, listed among the top 4 genes by geNorm and 3 top genes by NormFinder, was among two with the worst ranks according to Bestkeeper. A discrepancy between the results from BestKeeper and the results from both geNorm and NormFinder were reported in other studies (i.e.51,52). The reason for that is different input data (raw Cq values for BestKeeper and relative expression data for geNorm and NormFinder) and different stability criteria considered by the three programs – a matter thoroughly summarized in the works of De Spiegelaere et al.53 and Gomes et al.13.
Based on the juxtaposition of the results from geNorm, NormFinder and BestKeeper, we chose rho, rpoD and gyrB as the most promising RG candidates to be used for the normalization of RT-qPCR data in O. quorumnocens A44. Importantly, these genes do not belong to a common functional group (rho – encoding the transcription termination factor Rho, rpoD – encoding the primary sigma factor σ1, gyrB – encoding the B subunit of DNA gyrase1) what reduces the chances that they are co-regulated54,55.
Apart from ranking the potential RGs based on their stability, the geNorm tool also suggests the optimal number of genes to be included to the normalization factor46. In this study, based on the pilot assay, geNorm suggested including three RGs in gene expression studies in A44. This value, however, is a guideline and does not have to be treated as a strict threshold13,46. We investigated whether and to what extent the number of genes in the NF influences our expression data. The results showed moderate impact of switching form 2-gene (rho, gyrB) to 3-gene (rho, gyrB, rpoD) NF, with a difference observed only for a single out of 11 investigated culture conditions. However, application of a single-gene NF (rho) significantly influenced the outcome under 3/11 and 4/11 culture conditions when compared to 2-gen and 3-gene NFs, respectively. These results are in line with the MIQE guidelines, according to which expression of the target gene(s) should be normalized to more than one RG11.
The expression stability of rho, rpoD and gyrB, the most promising RG candidates selected in the pilot assay, was validated on the final dataset (11 culture conditions, all biological replicates). The geNorm M and CV values obtained for these genes were, respectively, M = 0.441 and CV = 0.187 for rho, M = 0.453 and CV = 0.185 for gyrB, and M = 0.5 and CV = 0.221 in case of rpoD. According to Hellemans and coworkers56, the recommended threshold stability values depend on the type of analyzed samples. For homogeneous samples, defined in medical studies as such originating from cell cultures of the same cell type, these requirements are more stringent and amount to M < 0.5 and CV < 0.25. For heterogeneous samples, in medical studies defined as originating from different cell types, clinical biopsies and cancer tissue in general, the criteria are less stringent (M < 1 and CV < 0.5). Transposition of these thresholds to prokaryotic studies is not straightforward. However, recent publications concerning RT-qPCR in prokaryotes suggest that for a given bacterial strain cultured in different conditions, genes with geNorm values M < 1 can be applied for normalization of RT-qPCR data10,13. In this study, rho, gyrB, rpoD met both aforementioned thresholds for acceptable stability.
In strain A44, the 16S rRNA-coding gene, a popular RG candidate in bacteria9, has shown poor expression stability and was not considered as a component of NF. In some studies, however, the target may meet the stability criteria (see Gomes et al., 201813). In such cases it should be considered that factors other than expression stability may also influence the performance of an RG. It was reported that while the turn-over of messenger RNAs in the cells is rather rapid, the ribosomal RNA is more stable and becomes degraded only under certain stress conditions57. These disproportions make a quantitative comparison between the two RNA types more difficult13. Another factor that comes to mind is the high disproportion in the Cq range observed between the potential genes of interest and the rRNA species, resulting from the high abundance of the latter in the cells. In RT-qPCR, however, as oppose to Northern blotting, it is not crucial for a good reference gene to be expressed at the same level as the gene of interest (Barbara D’haene, The gene expression blog, qbase+; https://blog.qbaseplus.com/four-tips-for-rt-qpcr-data-normalization-using-reference-genes).
A gfp-tagged derivative of the A44 strain44 was used throughout this study. Thus, apart from the genome-encoded RG candidates, we also investigated the stability of the heterologously expressed gfp. The gene is unique in terms of its occurrence in the terrestrial environment, therefore its stable expression in A44 GFP would create a low-cost alternative to the probe-based qPCR (i.e. TaqMan) for specific amplification of this RG candidate in complex samples. However, the stability of gfp in the tested setup proved insufficient. A potential cause may be uneven copy number per cell of the pPROBE-GTkan plasmid expressing gfp.
In this work, as a part of a broader study on the role of AiiO in the metabolism and environmental fitness of A44, we used RT-qPCR with the newly-established reference genes to investigate whether the expression of aiiO in A44 is altered upon exposure to AHLs and under a set of culture conditions. It is known that the expression of some enzyme-encoding genes may become highly elevated in the presence of cognate substrate(s), providing a solid link between a gene (protein) and a given compound or process. For example, Hommais et al.58 showed that the expression of pelD, a pectate lyase, can be induced over 10-fold in the stationary phase when polygalacturonate (substrate) is present.
The change in the expression of aiiO was investigated ninety minutes following the addition of synthetic AHLs (C6-HSL or 3OC12-HSL). The particular time point was chosen based on our previous experiments where the rate of exhaustion of AHLs following addition to cell culture was examined32. The consideration was to adjust the initial concentration of AHLs and time of sampling to prevent the total exhaustion of AHLs due to the activity of AiiO prior cell harvest.
Depending on the study, a gene is considered differentially regulated between two conditions when the change exceeds a 1-fold59,60 1.5-fold61,62 or a 2-fold threshold55. Using RT-qPCR, we showed that the expression of aiiO in O. quorumnocens A44 is not upregulated (fold change <159,60) upon supplementation with two synthetic AHLs, C6-HSL or 3OC12-HSL, nor in the presence of 3OC8-HSL and 3OC6-HSL and other compounds secreted to the medium by P. parmentieri SCC3193. No induction after addition of AHLs was reported earlier for aiiB and blcC genes encoding two QQ lactonases from A. tumefaciens42,63. Instead of being induced by signal molecules, the expression of those genes was enhanced in the presence of specific plant-derived compounds: succinic semialdehyde, gamma-hydroxybutyrate, gamma-butyrolactone, gamma-aminobutyrate and salicylic acid in case of blcC, and agrocinopine-enriched plant extracts in case of aiiB42,63,64. In this study, the expression of aiiO was investigated in the presence of water extract from potato roots. However, no significant up- or down-regulation of aiiO was observed in the applied setup.
Although the expression of aiiO was not distinctly upregulated in any of the conditions included in this study, we cannot exclude that such conditions exist. Some genes may be conserved in bacterial genomes, alike the aiiO homologues seem to be present in among different Ochrobactrum spp.32,65, even though they are useful only in a very particular niche or in the presence of certain environmental stress66. Ochrobactrum spp. are closely related to Brucella spp.14, known for their pathogenicity in a range of mammalian hosts67, and to Agrobacterium spp. and Rhizobium spp.15, known for establishing close interactions with plants68. Members of the Ochrobactrum genera were found in versatile environments, including soil, plants and the body of soil-dwelling organisms like the nematode Caenorhabditis elegans and the larvae of Holotrichia parallela69. Some Ochrobactrum spp. can cause opportunistic infections in humans, and one strain was proven to cause disease on mushrooms70. It is possible that the expression of aiiO becomes curtail (and therefore upregulated) in a very specific niche/host, upon certain abiotic stresses, or in the presence of some particular xenobiotic. Functional and structural similarities between the QQ enzymes and the xenobiotic/antibiotic-degrading enzymes were already denoted in several studies71,72.
In summary, we recommend rho, gyrB and rpoD as suitable RGs for gene expression analyses in O. quorumnocens A44. To our knowledge, this is the first study aimed to identify RGs for RT-qPCR analyses in a member of the Ochrobactrum genus. The expression of aiiO, a gene encoding the quorum quenching enzyme AiiO, was found not to be distinctly induced by the presence of AHLs, thereby providing no obvious indication that the primary substrate of AiiO are the AHLs.
Materials and Methods
Strains, chemicals and culture conditions
GFP-tagged derivative of O. quorumnocens A44 (A44 GFP)44, carrying plasmid pPROBE-GTkan45, was used throughout the experiments. The pVS1/p15a ori present on pPROBE-GTkan enables its stable propagation without selection73,74. The strain was grown under culture conditions listed in Table 1. Three types of basal media were used: Miller’s LB (Novagen, Germany), Miller’s LB agar (Novagen, Germany), and M63 mineral medium75. For the growth at pH 5.5, LB medium was buffered with 0.05M potassium hydrogen phthalate and the pH was adjusted to 5.5 with NaOH. To expose A44 GFP to metabolites secreted by the AHL-producing P. parmentieri SCC393176, the two strains were co-cultured in form of two parallel lines (approx. 4 mm wide), streaked on LB agar next to each other (~2 mm distance). The co-culture approach was adopted from plate experiments designed to screen bacterial strains for the production of AHLs using color-developing biosensor strains77. A44 GFP was also exposed to two synthetic AHLs: N-hexanoyl-L-homoserine lactone (C6-HSL) (Fluka/Sigma-Aldrich, USA) and N-3-oxo-dodecanoyl-L-homoserine lactone (3OC12-HSL) (Quorum Sensing Nottingham, UK). Two mL of an overnight culture of A44 GFP in M63 with 0.4% glucose was supplemented with 50 μM·mL−1 of one of the signal molecules and incubated for 90 minutes prior to cell harvest. The potato root extract was obtained from the roots of 3-week-old potato plants (cv. Dalia) grown in sandy soil under growth chamber conditions: 22 ± 1 °C temperature, 85 ± 5% relative humidity, 16/8 h light/dark photoperiod. The roots were harvested, washed with distilled water and homogenized in a filter bag (Bioreba AG, Switzerland) after addition of sterile water in 1:1 (w/v) ratio. The resulting 50% water extract was centrifuged (21000 RCF, 10′) and the supernatant was filter-sterilized (0.2 µm, cellulose acetate). For bacterial culture, the extract was mixed 1:1 with 2× concentrated M63 medium and supplemented with glucose to the final concentration of 0.4%. The experiments involving synthetic AHLs and potato root extract were performed in a defined mineral medium with a single carbon source to minimize the potential influence of compounds present in LB. To assess the expression of aiiO, a minimum of two biological replicates were carried out for each tested culture condition (Supplementary Table S3). The number of samples representing each condition is not even. No samples additionally processed for technical trials were arbitrary excluded from the dataset, resulting with uneven sample size between the analyzed treatments.
Selection of candidate RGs and primer design
The pool of candidate reference targets comprised 11 bacterial genes. Nine of them (16S rRNA, 23S rRNA, dnaK, groEL, gyrB, recA, rho, rpoB, rpoD) were chosen based on literature data. Homologues of these sequences were derived from the complete genome of O. quorumnocens A44 (CP022602.1–CP022605.1) (detailed list of loci in Table 2). The two other genes were: gfp, carried on the artificially introduced vector pPROBE-GTkan45 and CES85_4722, a hypothetical A44 gene with no data concerning its stability (a blind control). All PCR primers (Supplementary Table S1) were designed using the Primer3Plus software78,79 (http://primer3plus.com/). Specificity of the primers was verified using real-time PCR followed by melt curve analysis and electrophoresis in 1.2% agarose gel (TopVision, Thermo Scientific).
Isolation of RNA and cDNA synthesis
Total RNA from bacterial monocultures was isolated using RNeasy Protect Bacteria Mini Kit according to the manufacturer’s protocol (Qiagen, Gemany), with the optional on-column DNAse digestion. Approximately 4.5 × 108 cells were harvested and used per single isolation (cells harvested from 1 mL of cell suspension of turbidity equal to 5 units in McFarland’s scale). For cultures in liquid media, the turbidity was measured prior sampling and the volume of harvested cells was adjusted proportionally. Cells form solid media were suspended in sterile saline prior the turbidity measurement. The harvested cells (8000 RCF, 5′) were suspended in 500 μL of sterile saline to which 1 mL of RNA Protect Bacteria reagent (Qiagen, Gemany) was immediately added to prevent RNA decay.
All RNA samples were treated with TURBO DNA-free Kit (Thermo Scientific, USA) to remove any gDNA contamination and stored at −80 °C. The concentration and the quality of RNA for gene expression stability assessment was evaluated using an Agilent 2100 Bioanalyzer Instrument and an Agilent RNA 6000 Nano Kit (Agilent Technologies, USA). All processed samples had RNA Integrity value (RIN) ≥ 8.7. RIN values between 8–10 indicate intact, high quality RNA samples and RIN = 5.0 is the suggested minimum threshold for applicability of the given RNA in downstream RT-qPCR analyses80,81. Next, RNA was reverse-transcribed to cDNA with Transcriptor First Strand cDNA Synthesis Kit (Roche, Poland) using random hexamer primers. The optional denaturation step was included. Total amount of RNA used per reaction was 500 ng.
qPCR
The assays were carried out using the CFX96 instrument (Bio-Rad), in a 96-well plate format. Reaction mixtures of 20 μL included: 2 × Power SYBR Green Mastermix (Thermo Scientific, USA), forward and reverse primers at a final concentration of 300 nM and 4 μL of diluted (1:3) post-RT mixture containing cDNA. The PCR conditions were 95 °C for 10 min, 40 cycles of 95 °C for 15S and 60 °C for 1 min, with a final melt curve 55–95 °C at a 0.5 °C/5S increment. Each reaction was performed in duplicate. The difference between duplicates was <0.5 cycle for 98% of reactions. Inter-run calibrator (IRC) was included in each subsequent plate. Parameters of the assay, including sensitivity, linearity and primer efficiency, were validated using a 6-point 10-fold serial dilution of templates, for which the corresponding post-PCR amplicons were applied.
Expression stability and the optimal number of RGs
Eleven candidate RGs were ranked according to their expression stability under 10 culture conditions. Initially, the expression stability was evaluated in a geNorm46 pilot assay where a set of 10 cDNA samples (one per condition) was targeted for all RG candidates. geNorm incorporated into the qbase+ software, version 2.3 (Biogazelle NV, Belgium) was applied. In a geNorm pilot study, typically 8–10 candidate RGs are measured in a set of samples (usually 10). It is recommended that no sample type is overrepresented, what could introduce bias in terms of candidate genes being more stable in a given condition55. The resulting values (geNorm M) were compared with RG ranks obtained with NormFinder47 and BestKeeper48. Each of the applied methods is based on a different statistical approach, therefore the obtained results are not identical. To make our decision on the choice of a specific approach less arbitrary, we conducted a juxtaposition of the results obtained using the three algorithms by calculating a geometric mean. Genes with the best overall score were selected for further experiments. The expression stability the RGs pre-selected in the pilot was verified on the final dataset (11 culture conditions, 2–3 biological replicates) – the same one used to investigate the expression of aiiO. The validation was conducted using geNorm (M values).
The optimal number of RGs to be included in the normalization factor was established by calculating the Vn/Vn + 1 pair-wise variation, where n is the number of RGs in the NF46. In this method, a V value below 0.15 indicates no benefit of going from n to n + 1.
The performance of different normalization factors was compared on CNRQ values where the expression of aiiO was normalized to either one (NF1; rho), two (NF2; rho, gyrB) or three (NF3; rho, gyrB, rpoD) RGs. The statistical significance (α = 0.05) of pair-wise differences between groups normalized with different NFs was calculated using two-tailed Student’s t-test with unequal variation (Welch’s t-test).
Analysis of gene expression
Gene expression analysis was conducted using the qbase+ software, version 2.3 (Biogazelle NV, Belgium). Subsequent steps were involved: the relative quantity (RQ) was calculated using ∆∆Ct method82, the RQ was normalized with respect to the established RGs (NRQ) and the value was calibrated with respect to the applied inter-run calibrator samples to obtain calibrated NRQ (CNRQ) interpreted as the relative expression. The significance of changes in the expression of aiiO between the ‘control’ and the ‘treated’ samples was evaluated using the BootstRatio tool for statistical analysis of fold-change (http://pdo.iconcologia.net/stats/br/index.html)83. To plot the fold change, CNRQ values were log base 2 transformed with respect to median of the respective reference samples for the three tested media types (LB agar, LB, M63 0.4% glucose).
Supplementary information
Acknowledgements
The authors thank R. Czajkowski (IFB UG&MUG, Gdańsk) for valuable editorial comments to the manuscript. This research was funded by the National Science Centre, Poland (Narodowe Centrum Nauki, Polska) research grant OPUS7 no. 2014/13/B/NZ9/02136 to S.J.; D.M.K. was supported by a personal scholarship START 2017 (START 40.2017) granted by the Foundation for Polish Science (Fundacja na Rzecz Nauki Polskiej, FNP).
Author Contributions
Conceptualization: D.M.K. and S.J.; Methodology: A.S. and D.K.; Investigation: A.S., D.M.K., T.M., M.M.; Writing-Original Draft Preparation: D.M.K.; Writing-Review & Editing, S.J., D.M.K.; Visualization, D.M.K.; Supervision, S.J.; Funding Acquisition, S.J.
Data Availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-49474-6.
References
- 1.Galisa PS, et al. Identification and validation of reference genes to study the gene expression in Gluconacetobacter diazotrophicus grown in different carbon sources using RT-qPCR. J. Microbiol. Methods. 2012;91:1–7. doi: 10.1016/j.mimet.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Menna P, Barcellos FG, Hungria M. Phylogeny and taxonomy of a diverse collection of Bradyrhizobium strains based on multilocus sequence analysis of the 16S rRNA gene, ITS region and glnII, recA, atpD and dnaK genes. Int. J. Syst. Evol. Microbiol. 2009;59:2934–50. doi: 10.1099/ijs.0.009779-0. [DOI] [PubMed] [Google Scholar]
- 3.Florindo C, et al. Selection of reference genes for real-time expression studies in Streptococcus agalactiae. J. Microbiol. Methods. 2012;90:220–227. doi: 10.1016/j.mimet.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Bhubhanil S, Niamyim P, Sukchawalit R, Mongkolsuk S. Cysteine desulphurase-encoding gene sufS2 is required for the repressor function of RirA and oxidative resistance in Agrobacterium tumefaciens. Microbiology. 2014;160:79–90. doi: 10.1099/mic.0.068643-0. [DOI] [PubMed] [Google Scholar]
- 5.Mirabella A, et al. Brucella melitensis MucR, an orthologue of Sinorhizobium meliloti MucR, is involved in resistance to oxidative, detergent, and saline stresses and cell envelope modifications. J. Bacteriol. 2013;195:453–65. doi: 10.1128/JB.01336-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliveira LR, Rodrigues EP, Marcelino-Guimarães FC, Oliveira ALM, Hungria M. Fast induction of biosynthetic polysaccharide genes lpxA, lpxE, and rkpI of Rhizobium sp. strain PRF 81 by common bean seed exudates is indicative of a key role in symbiosis. Funct. Integr. Genomics. 2013;13:275–83. doi: 10.1007/s10142-013-0322-7. [DOI] [PubMed] [Google Scholar]
- 7.Cleenwerck I, De Vos P, De Vuyst L. Phylogeny and differentiation of species of the genus Gluconacetobacter and related taxa based on multilocus sequence analyses of housekeeping genes and reclassification of Acetobacter xylinus subsp. Sucrofermentans as Gluconacetobacter sucrofermentans (T. Int. J. Syst. Evol. Microbiol. 2010;60:2277–83. doi: 10.1099/ijs.0.018465-0. [DOI] [PubMed] [Google Scholar]
- 8.Stenico V, Baffoni L, Gaggìa F, Biavati B. Validation of candidate reference genes in Bifidobacterium adolescentis for gene expression normalization. Anaerobe. 2014;27:34–39. doi: 10.1016/j.anaerobe.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Rocha DJP, Santos CS, Pacheco LGC. Bacterial reference genes for gene expression studies by RT-qPCR: survey and analysis. Antonie Van Leeuwenhoek. 2015;108:685–693. doi: 10.1007/s10482-015-0524-1. [DOI] [PubMed] [Google Scholar]
- 10.Kałużna M, Kuras A, Puławska J. Validation of reference genes for the normalization of the RT-qPCR gene expression of virulence genes of Erwinia amylovora in apple shoots. Sci. Rep. 2017;7:2034. doi: 10.1038/s41598-017-02078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bustin SA, et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 12.DeLorenzo DM, Moon TS. Selection of stable reference genes for RT-qPCR in Rhodococcus opacus PD630. Sci. Rep. 2018;8:6019. doi: 10.1038/s41598-018-24486-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomes AÉI, et al. Selection and validation of reference genes for gene expression studies in Klebsiella pneumoniae using Reverse Transcription Quantitative real-time PCR. Sci. Rep. 2018;8:9001. doi: 10.1038/s41598-018-27420-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velasco J, et al. Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov., a new species with a closer relationship to Brucella spp. Int. J. Syst. Bacteriol. 1998;48:759–768. doi: 10.1099/00207713-48-3-759. [DOI] [PubMed] [Google Scholar]
- 15.Scholz HC, et al. Genetic diversity and phylogenetic relationships of bacteria belonging to the Ochrobactrum–Brucella group by recA and 16S rRNA gene-based comparative sequence analysis. Syst. Appl. Microbiol. 2008;31:1–16. doi: 10.1016/j.syapm.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Bohlin J, et al. Genomic comparisons of Brucella spp. and closely related bacteria using base compositional and proteome based methods. BMC Evol. Biol. 2010;10:249. doi: 10.1186/1471-2148-10-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes B, Popoff M, Kiredjian M, Kersters K. Ochrobactrum anthropi gen. nov., sp. nov. from Human Clinical Specimens and Previously Known as Group Vd. Int. J. Syst. Bacteriol. 1988;38:406–416. doi: 10.1099/00207713-38-4-406. [DOI] [Google Scholar]
- 18.Trujillo ME, et al. Nodulation of Lupinus albus by strains of Ochrobactrum lupini sp. nov. Appl. Environ. Microbiol. 2005;71:1318–27. doi: 10.1128/AEM.71.3.1318-1327.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zurdo-Piñeiro JL, et al. Ochrobactrum cytisi sp. nov., isolated from nodules of Cytisus scoparius in Spain. Int. J. Syst. Evol. Microbiol. 2007;57:784–8. doi: 10.1099/ijs.0.64613-0. [DOI] [PubMed] [Google Scholar]
- 20.Li L, et al. Ochrobactrum endophyticum sp. nov., isolated from roots of Glycyrrhiza uralensis. Arch. Microbiol. 2016;198:171–179. doi: 10.1007/s00203-015-1170-8. [DOI] [PubMed] [Google Scholar]
- 21.Kampfer P, et al. Ochrobactrum rhizosphaerae sp. nov. and Ochrobactrum thiophenivorans sp. nov., isolated from the environment. Int. J. Syst. Evol. Microbiol. 2008;58:1426–1431. doi: 10.1099/ijs.0.65407-0. [DOI] [PubMed] [Google Scholar]
- 22.Dirksen P, et al. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol. 2016;14:38. doi: 10.1186/s12915-016-0258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu X-H, et al. Isolation and characterization of a bacterial strain of the genus Ochrobactrum with methyl parathion mineralizing activity. J. Appl. Microbiol. 2006;101:986–994. doi: 10.1111/j.1365-2672.2006.03016.x. [DOI] [PubMed] [Google Scholar]
- 24.Woźniak-Karczewska, M. et al. Isolation of two Ochrobactrum sp. strains capable of degrading the nootropic drug—Piracetam. N. Biotechnol. (2017). [DOI] [PubMed]
- 25.Sanjeev Kumar S, Kumar MS, Siddavattam D, Karegoudar TB. Generation of continuous packed bed reactor with PVA–alginate blend immobilized Ochrobactrum sp. DGVK1 cells for effective removal of N,N-dimethylformamide from industrial effluents. J. Hazard. Mater. 2012;199–200:58–63. doi: 10.1016/j.jhazmat.2011.10.053. [DOI] [PubMed] [Google Scholar]
- 26.Sipahutar MK, Vangnai AS. Role of plant growth-promoting Ochrobactrum sp. MC22 on triclocarban degradation and toxicity mitigation to legume plants. J. Hazard. Mater. 2017;329:38–48. doi: 10.1016/j.jhazmat.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 27.Sumayo M, Hahm M-S, Ghim S-Y. Determinants of plant growth-promoting Ochrobactrum lupini KUDC1013 involved in induction of systemic resistance against Pectobacterium carotovorum subsp. carotovorum in tobacco leaves. Plant Pathol. J. 2013;29:174–181. doi: 10.5423/PPJ.SI.09.2012.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulucci NS, et al. Arachis hypogaea PGPR isolated from Argentine soil modifies its lipids components in response to temperature and salinity. Microbiol. Res. 2015;173:1–9. doi: 10.1016/j.micres.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 29.Chakraborty U, Chakraborty BN, Basnet M, Chakraborty AP. Evaluation of Ochrobactrum anthropi TRS-2 and its talc based formulation for enhancement of growth of tea plants and management of brown root rot disease. J. Appl. Microbiol. 2009;107:625–34. doi: 10.1111/j.1365-2672.2009.04242.x. [DOI] [PubMed] [Google Scholar]
- 30.Krzyżanowska DM, et al. Ochrobactrum quorumnocens sp. nov., a quorum quenching bacterium from the potato rhizosphere, and comparative genome analysis with related type strains. PLoS One. 2019;14:e0210874. doi: 10.1371/journal.pone.0210874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jafra S, et al. Detection and characterization of bacteria from the potato rhizosphere degrading N-acyl-homoserine lactone. Can. J. Microbiol. 2006;52:1006–15. doi: 10.1139/w06-062. [DOI] [PubMed] [Google Scholar]
- 32.Czajkowski R, et al. Inactivation of AHLs by Ochrobactrum sp. A44 depends on the activity of a novel class of AHL acylase. Environ. Microbiol. Rep. 2011;3:59–68. doi: 10.1111/j.1758-2229.2010.00188.x. [DOI] [PubMed] [Google Scholar]
- 33.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994;176:269–75. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papenfort K, Bassler BL. Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016;14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong YH, et al. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature. 2001;411:813–7. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- 36.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 2009;7:836–836. doi: 10.1038/nrmicro2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh RP. Attenuation of quorum sensing-mediated virulence in Gram-negative pathogenic bacteria: implications for the post-antibiotic era. Med. Chem. Commun. 2015;6:259–272. doi: 10.1039/C4MD00363B. [DOI] [Google Scholar]
- 38.Pirhonen M, Flego D, Heikinheimo R, Palva ET. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 1993;12:2467–76. doi: 10.1002/j.1460-2075.1993.tb05901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Czajkowski R, Jafra S. Quenching of acyl-homoserine lactone-dependent quorum sensing by enzymatic disruption of signal molecules. Acta Biochim. Pol. 2009;56:1–16. doi: 10.18388/abp.2009_2512. [DOI] [PubMed] [Google Scholar]
- 40.Grandclément C, Tannières M, Moréra S, Dessaux Y, Faure D. Quorum quenching: role in nature and applied developments. FEMS Microbiol. Rev. 2016;40:86–116. doi: 10.1093/femsre/fuv038. [DOI] [PubMed] [Google Scholar]
- 41.Roche DM, et al. Communications blackout? Do N-acylhomoserine-lactone-degrading enzymes have any role in quorum sensing? Microbiology. 2004;150:2023–8. doi: 10.1099/mic.0.26977-0. [DOI] [PubMed] [Google Scholar]
- 42.Khan SR, Farrand SK. The BlcC (AttM) lactonase of Agrobacterium tumefaciens does not quench the quorum-sensing system that regulates Ti plasmid conjugative transfer. J. Bacteriol. 2009;191:1320–9. doi: 10.1128/JB.01304-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nadal Jimenez P, et al. Role of PvdQ in Pseudomonas aeruginosa virulence under iron-limiting conditions. Microbiology. 2010;156:49–59. doi: 10.1099/mic.0.030973-0. [DOI] [PubMed] [Google Scholar]
- 44.Krzyzanowska DM, Obuchowski M, Bikowski M, Rychłowski M, Jafra S. Colonization of potato rhizosphere by GFP-tagged Bacillus subtilis MB73/2, Pseudomonas sp. P482 and Ochrobactrum sp. A44 shown on large sections of roots using enrichment sample preparation and confocal laser scanning microscopy. Sensors (Basel). 2012;12:17608–19. doi: 10.3390/s121217608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller WG, Leveau JH, Lindow SE. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant. Microbe. Interact. 2000;13:1243–50. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- 46.Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034 (2002). [DOI] [PMC free article] [PubMed]
- 47.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 48.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 49.Taylor S, Wakem M, Dijkman G, Alsarraj M, Nguyen M. A practical approach to RT-qPCR—Publishing data that conform to the MIQE guidelines. Methods. 2010;50:S1–S5. doi: 10.1016/j.ymeth.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 50.Supernat A, et al. Epithelial-mesenchymal transition and cancer stem cells in endometrial cancer. Anticancer Res. 2013;33:5461–9. [PubMed] [Google Scholar]
- 51.Martínez-Giner M, Noguera JL, Balcells I, Fernández-Rodríguez A, Pena RN. Selection of internal control genes for real-time quantitative PCR in ovary and uterus of sows across pregnancy. PLoS One. 2013;8:e66023. doi: 10.1371/journal.pone.0066023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robledo D, et al. Analysis of qPCR reference gene stability determination methods and a practical approach for efficiency calculation on a turbot (Scophthalmus maximus) gonad dataset. BMC Genomics. 2014;15:648. doi: 10.1186/1471-2164-15-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Spiegelaere W, et al. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS One. 2015;10:e0122515. doi: 10.1371/journal.pone.0122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silver N, Best S, Jiang J, Thein S. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hellemans, J. & Vandesompele, J. qPCR data analysis – unlocking the secret to successful results. PCR troubleshooting and optimization: the essential guide, 139–150 (2011).
- 56.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–66. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hommais F, et al. lpxC and yafS are the most suitable internal controls to normalize real time RT-qPCR expression in the phytopathogenic bacteria Dickeya dadantii. PLoS One. 2011;6:e20269. doi: 10.1371/journal.pone.0020269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeller T, Moskvin OV, Li K, Klug G, Gomelsky M. Transcriptome and physiological responses to hydrogen peroxide of the facultatively phototrophic bacterium Rhodobacter sphaeroides. J. Bacteriol. 2005;187:7232–42. doi: 10.1128/JB.187.21.7232-7242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Varano M, et al. Temperature-dependent regulation of the Ochrobactrum anthropi proteome. Proteomics. 2016;16:3019–3024. doi: 10.1002/pmic.201600048. [DOI] [PubMed] [Google Scholar]
- 61.Puławska J, Kałużna M, Warabieda W, Mikiciński A. Comparative transcriptome analysis of a lowly virulent strain of Erwinia amylovora in shoots of two apple cultivars – susceptible and resistant to fire blight. BMC Genomics. 2017;18:868. doi: 10.1186/s12864-017-4251-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fan B, et al. Transcriptomic profiling of Bacillus amyloliquefaciens FZB42 in response to maize root exudates. BMC Microbiol. 2012;12:116. doi: 10.1186/1471-2180-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haudecoeur E, et al. Different Regulation and Roles of Lactonases AiiB and AttM in Agrobacterium tumefaciens C58. Mol. Plant-Microbe Interact. 2009;22:529–537. doi: 10.1094/MPMI-22-5-0529. [DOI] [PubMed] [Google Scholar]
- 64.Lang J, Faure D. Functions and regulation of quorum-sensing in Agrobacterium tumefaciens. Front. Plant Sci. 2014;5:14. doi: 10.3389/fpls.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mei G-Y, Yan X-X, Turak A, Luo Z-Q, Zhang L-Q. AidH, an alpha/beta-hydrolase fold family member from an Ochrobactrum sp. strain, is a novel N-acylhomoserine lactonase. Appl. Environ. Microbiol. 2010;76:4933–42. doi: 10.1128/AEM.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rohmer L, Hocquet D, Miller SI. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 2011;19:341–8. doi: 10.1016/j.tim.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whatmore AM. Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect. Genet. Evol. 2009;9:1168–1184. doi: 10.1016/j.meegid.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 68.Gnanamanickam, S. S. Plant-Associated Bacteria. (Springer Netherlands, 2006).
- 69.Huang S, Sheng P, Zhang H. Isolation and identification of cellulolytic bacteria from the gut of Holotrichia parallela larvae (Coleoptera: Scarabaeidae) Int. J. Mol. Sci. 2012;13:2563–2577. doi: 10.3390/ijms13032563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu Z, et al. Mushroom tumor: a new disease on Flammulina velutipes caused by Ochrobactrum pseudogrignonense. FEMS Microbiol. Lett. 2016;363:fnv226. doi: 10.1093/femsle/fnv226. [DOI] [PubMed] [Google Scholar]
- 71.Elias M, Tawfik DS. Divergence and convergence in enzyme evolution: parallel evolution of paraoxonases from quorum-quenching lactonases. J. Biol. Chem. 2012;287:11–20. doi: 10.1074/jbc.R111.257329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tannières M, et al. A Metagenomic study highlights phylogenetic proximity of quorum-quenching and xenobiotic-degrading amidases of the AS-family. PLoS One. 2013;8:65473. doi: 10.1371/journal.pone.0065473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hallmann J, Quadt-Hallmann A, Miller WG, Sikora RA, Lindow SE. Endophytic Colonization of Plants by the Biocontrol Agent Rhizobium etli G12 in Relation to Meloidogyne incognita Infection. Phytopathology. 2001;91:415–422. doi: 10.1094/PHYTO.2001.91.4.415. [DOI] [PubMed] [Google Scholar]
- 74.Heeb S, et al. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in Gram-negative, plant-associated bacteria. Mol. Plant-Microbe Interact. 2000;13:232–237. doi: 10.1094/MPMI.2000.13.2.232. [DOI] [PubMed] [Google Scholar]
- 75.Miller, J. H. Experiments in molecular genetics. (Cold Spring Harbor Laboratory, 1972).
- 76.Pirhonen M, Saarilahti H, Karlsoon MB, Palva ET. Identification of pathogenicity determinants of Erwinia carotovora subsp. carotovora by transposon mutagenesis. Mol. Plant Microbe Interact. 1991;4:276–283. doi: 10.1094/MPMI-4-276. [DOI] [Google Scholar]
- 77.Ravn L, Christensen AB, Molin S, Givskov M, Gram L. Methods for detecting acylated homoserine lactones produced by Gram-negative bacteria and their application in studies of AHL-production kinetics. J. Microbiol. Methods. 2001;44:239–251. doi: 10.1016/S0167-7012(01)00217-2. [DOI] [PubMed] [Google Scholar]
- 78.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–91. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 79.Untergasser A, et al. Primer3 – new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 81.Pazzagli M, et al. SPIDIA-RNA: First external quality assessment for the pre-analytical phase of blood samples used for RNA based analyses. Methods. 2013;59:20–31. doi: 10.1016/j.ymeth.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 82.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 83.Clèries R, et al. BootstRatio: A web-based statistical analysis of fold-change in qPCR and RT-qPCR data using resampling methods. Comput. Biol. Med. 2012;42:438–445. doi: 10.1016/j.compbiomed.2011.12.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).