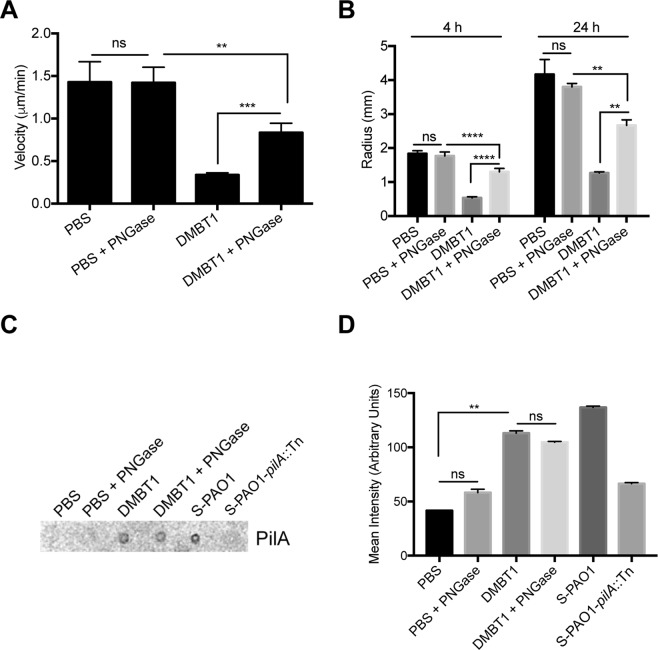
Figure 8.
DMBT1 inhibition of P. aeruginosa twitching motility involves its N-Glycosylation. DMBT1 (100 ng/µL) was digested by PNGase F (glycerol-free) at 37 °C for 2 days in native glycobuffer (50 mM sodium phosphate, pH 7.5) to remove N-glycosylation of DMBT1. PBS and DMBT1 diluted with same volume of native glycobuffer, and PBS treated with same volume of PNGase served as controls. (A) Effect of DMBT1 digested by PNGase F on twitching velocity of P. aeruginosa PAO1 after 4 h. (B) Effects of DMBT1 digested by PNGase F on colony size of P. aeruginosa PAO1 after 4 h and 24 h. Data are shown as the mean ± SEM per sample from three independent experiments. Significance was determined using one-way ANOVA (panel A) or two-way ANOVA (panel B) each with Tukey’s multiple comparisons, ****P < 0.0001; ***P < 0.001; **P < 0.01; ns, not significant. (C) Dot-immunoblot assay using anti-PilA antibody showing the binding of PAO1 pili to purified DMBT1 (400 ng) even after PNGase F digestion. Pili extracts from PAO1 (S-PAO1), extracts from the pilA mutant (S-PAO1-pilA::Tn), both diluted 1 in 500 in PBS were used as controls, along with PBS alone. (D) Quantification of dot-intensity using ImageJ from the dot-immunoblot assay shown in panel C. Data are shown as the mean ± SEM of duplicate measurements from each sample. Significance determined used Kruskal-Wallis test with Dunn’s multiple comparisons, **P < 0.01; ns, not significant.