Figure 3.
The Mitochondrial Redox Balance Is Significantly Altered after 50 mGy of LDIR in Primary Mouse Keratinocytes
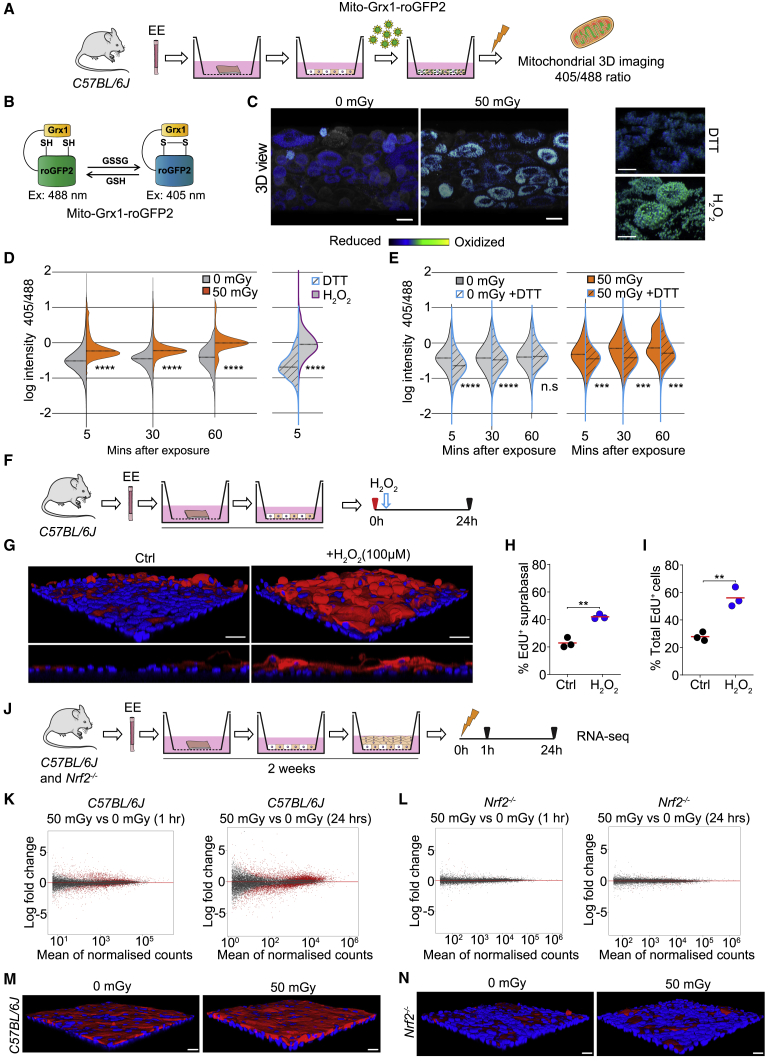
(A) Experimental protocol. Primary 3D cultures of EE were infected with an adenovirus encoding a genetic sensor of the mitochondrial redox state, irradiated, and imaged.
(B) The Mito-Grx1-roGFP2 reporter is localized to mitochondria. The reduced and oxidized states of the probe are differentially excited by 405 nm and 488 nm light, so the ratio of fluorescence after excitation at the two wavelengths indicates the redox state (Gutscher et al., 2008).
(C) Representative rendered confocal z stacks showing 405 nm/488 nm emission ratios from mitochondria in Mito-Grx1-roGFP2-expressing keratinocytes 60 min after 0 or 50 mGy LDIR, indicated by the pseudo-color scale. Scale bars, 20 μm. Negative (DTT-treated) and positive (H2O2-treated) controls for oxidation are shown as well. Scale bars, 15 μm.
(D) Violin plots of the distribution of 405/488 ratios for individual mitochondria in Mito-Grx1-roGFP2 reporter-expressing keratinocytes 5, 30, and 60 min after LDIR, obtained by quantitative confocal 3D imaging. Controls are oxidized (hydrogen peroxide [H2O2]-treated) and reduced (DTT-treated) cells ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001 (Mann-Whitney U test). n is the number of mitochondria imaged under each condition, shown in Table S2. Three biological replicate experiments were performed; results from a representative experiment are shown.
(E) Violin plots showing the effect of DTT treatment on irradiated cells. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001 (Mann-Whitney U test).
(F–I) Experimental protocol.
(F) Primary 3D cultures of EE were labeled with EdU for 1 h and treated with H2O2 24 h before immunostaining.
(G) Rendered confocal z stacks of typical cultures 24 h after treatment with the control (Ctrl) or 100 μM H2O2. Differentiated suprabasal keratinocytes were stained for KRT4 (red) and DAPI (blue). Dashed lines indicate the insert’s membrane. Scale bars, 20 μm.
(H and I) In vitro EdU lineage tracing. Shown are the percentage of EdU+ suprabasal cells (H) and percentage of EdU+ total cells (I) after treatment with the Ctrl or 100 μM H2O2. Each point represents the mean from a biological replicate culture from a different mouse. ∗∗p < 0.01 (unpaired t test), n = 3; total EdU+ cells, 2,435 (Ctrl) and 3,256 (H2O2).
(J–L) Transcriptional profile of irradiated EE cultures from wild-type C57BL/6J and Nrf2−/− mice.
(J) Experimental protocol. RNA-seq was performed on biological triplicate cultures 1 and 24 h after 0 or 50 mGy of LDIR.
(K and L) MA plots of RNA-seq data of cultures from C57BL/6J (K) and Nrf2−/− mice (L) comparing irradiated and unirradiated cultures at the times shown; red indicates differentially expressed transcripts with adjusted p < 0.05.
(M and N) Rendered confocal z stacks of C57BL/6J (M) and Nrf2−/− (N) cultures 24 h after 0 or 50 mGy LDIR. Differentiated suprabasal keratinocytes were stained for KRT4 (red) and DAPI (blue). Scale bars, 25 μm.