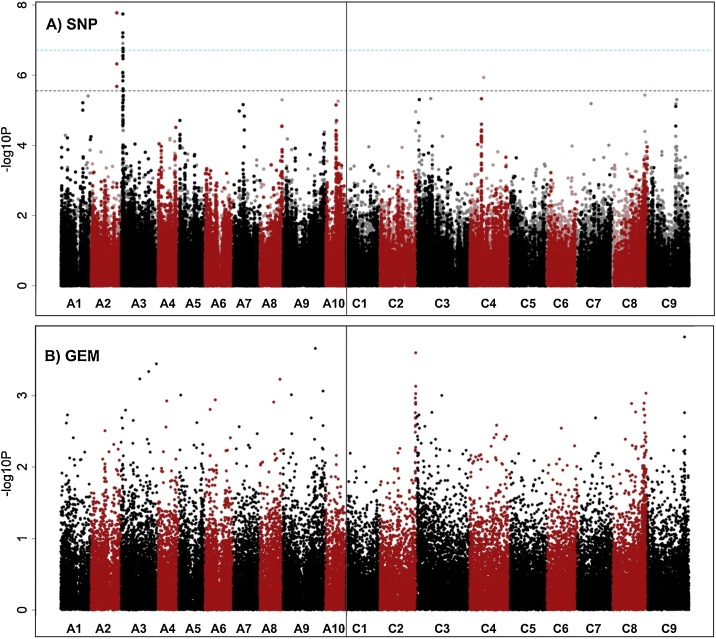
Fig. 5.
Association analysis for root aromatic glucosinolate content. (A) Manhattan plot showing genome-wide associations for the identification of transcriptome single-nucleotide polymorphism (SNP) markers of 288 Brassica napus accessions with leaf glucosinolate content. Marker associations was calculated using a mixed linear model which incorporated population structure and relatedness. The SNP markers are positioned on the x-axis based on the genomic order of the gene models in which the polymorphism was scored. The significance of the trait association, as -log10 P values, plotted on the y-axis. The horizontal purple and cyan lines represent false discovery rate (FDR) threshold at 5% and the threshold for Bonferroni significance of 0.05, respectively. Chromosomes of B. napus are labelled A1– A10 and C1 – C9, shown in alternating black and red colors to allow boundaries to be clearly distinguished. Dark opaque points are simple SNP markers (i.e. polymorphisms between resolved bases) and hemi-SNPs that have been directly linkage-mapped, both of which can be assigned to one genome, whereas light points are hemi-SNP markers (i.e. polymorphisms involving multiple bases called at the SNP position in one allele of the polymorphism) for which the genome of the polymorphism cannot be assigned. (B) Association analysis of expression variation-based markers (GEM) with leaf aliphatic glucosinolate. Reads per kb per million aligned reads (RPKM) were regressed against the trait, and R2 and P values were calculated for each gene. The gene models are positioned on the x-axis based on their genomic order, with the significance of the associated trait, as -log10 P, plotted on the y-axis (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).