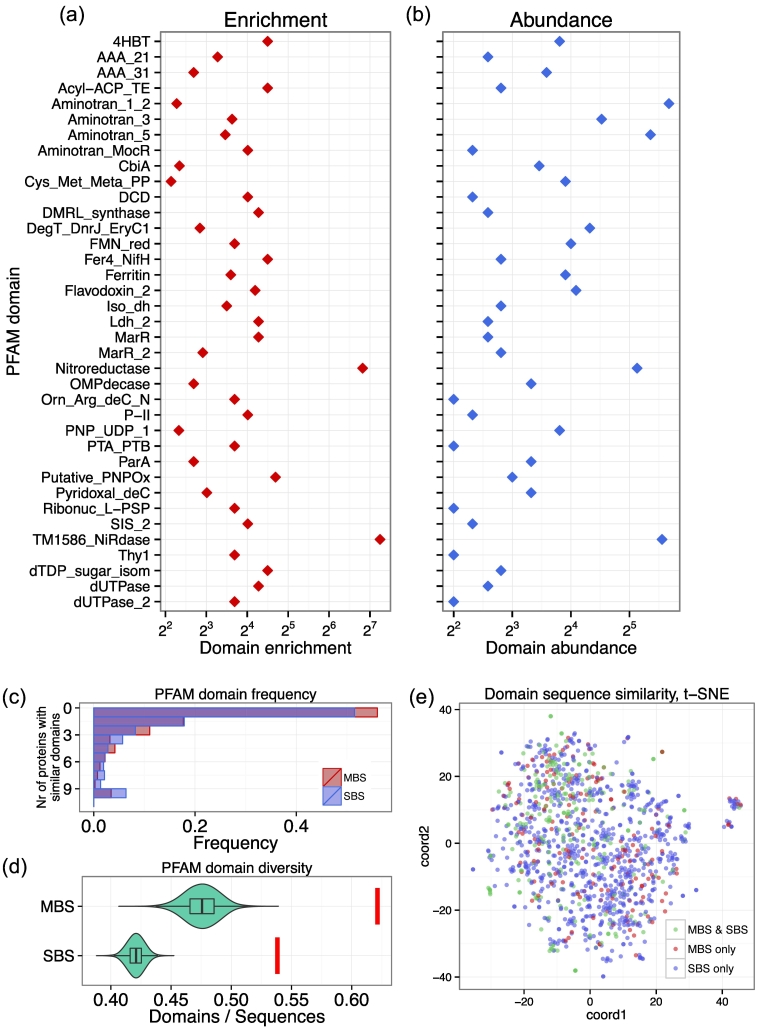
Fig. 4.
Characteristics of conserved domains in MBS and SBS homomers with a single PFAM domain. (A) PFAM domains that are significantly enriched in MBS homomers (FDR-corrected p < 0.05) compared to SBS homomers. For plotting, in the case of domains that are absent in SBS homomers, we used 1 as their abundance in SBS homomers (but not for significance testing). (B) The abundance of significantly enriched PFAM domains in MBS homomers. (C) The frequency distribution of PFAM domains does not differ qualitatively in MBS and SBS homomers: in both cases, more than 50% of domains are present in only a single protein, and ~ 80% of domains are present in three or less proteins. Note that domains present in more than 10 proteins are included in bin with 10, and domain overlaps (i.e., more than one different domains mapping to the same sequence region) were permitted. (D) MBS homomers have a somewhat higher observed diversity (0.62, red vertical bar) of PFAM domains than SBS homomers (0.54). Similarly, their expected diversity obtained through Monte-Carlo simulations (violin plots) is higher than in SBS homomers. Diversity was measured as the number of domains per the number of sequences. (E) Two-dimensional (t-SNE) representation of the sequence space of PFAM domains of homomers. Neither domains specific for MBS or SBS homomers nor domains present in both of them form separate clusters, but are distributed relatively evenly across the sequence space. (See also Fig. S4 for their full sequences.) Taken together, these results indicate that the differences between MBs and SBS homomers are not caused by the more limited diversity of domains in MBS homomers.