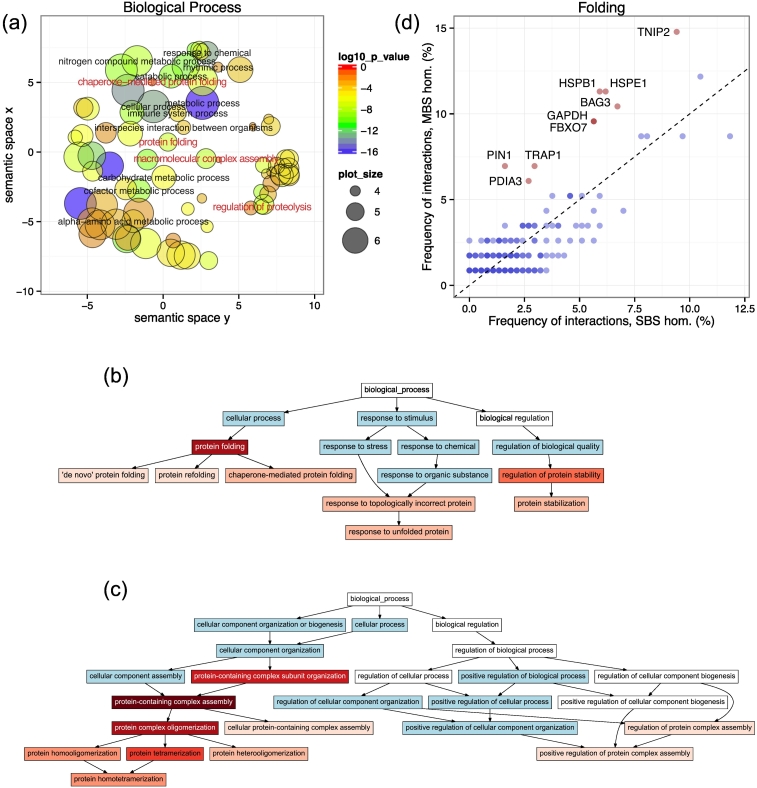
Fig. 5.
Folding, assembly and degradation related GO terms are enriched in the interactome of human MBS homomers. (A) Summary (made with REVIGO) of the main categories of the Biological Process GO terms that are significantly enriched in the interactors of MBS homomers compared to the interactors of all homomers (MBS + SBS). Altogether, more than 600 terms are significantly enriched, many of them related to protein folding, assembly and proteolysis (highlighted with red). These include “chaperone mediated protein folding,” “protein folding,” “macromolecular complex assembly” and “regulation of proteolysis.” (see supplementary data for the full GO enrichment results). (B) Graph of the significantly enriched terms related to protein folding and protein stabilization. The intensity of red corresponds to significance, while terms in blue are also (highly) significant but are too high level to be considered as folding related. (C) Graph of the significantly enriched terms related to assembly of protein complexes. Color coding is similar to panel B. (D) Frequencies of interactions of folding related genes in MBS and SBs homomers. A gene with x = 4% and y = 5% interacts with 4% of SBS homomers and 5% of MBS homomers. Most genes fall close to the x = y line, that is, have a similar frequency of interactions in MBS homomers and SBS homomers; however, nine genes form a separate cluster (highlighted with red) and are more distant from the parity line than 3 standard deviations of the remaining genes. These genes include an HSP70/HSC70 chaperone regulator BAG3, a small chaperone protein HSPB1, a co-chaperonin HSPE1 and a 75 kDa heat shock protein TRAP1 (see also Supplementary Table 3).